Lupine Publishers | Current Investigations in Agriculture and Current Research
Abstract
This study examined Information and Communication Technology (ICTs) roles in improving women farmer’s access to agricultural services and agribusiness in Orlu Agricultural Zone of Imo State. Data were collected with structured questionnaire distributed to 110 respondents randomly selected from the 10 extension blocks in Orlu Agricultural zone. Data collected were analyzed using percentages, mean scores and standard deviation. The results obtained showed that ICT devices available in the area included radio, mobile phones, television and newspaper among others. However, mobile phone is the most readily available (ICT) device (87.3%), followed by Radio (74.5%). It was also observed that ICTs play significant roles in improving women’s access to agricultural service and agribusiness as as indicated by a mean score of 2.50 Several factors constrained the use of ICT devices. Among these factors were; network problems/connectivity(M=4.00), high cost of ICT devices (M=3.47), widespread illiteracy (M = 2.96), low level of awareness (M=2.55), poor saving ability (M=2.47), insufficient income (M=2.12), and language illiteracy (M=2.00). This study thus recommended that government should provide rural infrastructure especially electricity supply, good road network etc in the study area, establishment of ICT centers where farmers can acquire practical computer training for the enhancement of ICT use. Information on agricultural services and agribusiness should be made available to farmers through their mobile phones in their local language and it should be timely for effective utilization.
Keywords: Agriculture; Agribusiness; ICTs; Farmers; Services; Market; Women
Introduction
Globally, agriculture takes center stage as the engine that can transform nations’ economies. Similarly, the place and role of smallholder farmers from local to global levels are recognized by the respective governments and international partners as a way to avoid age-old problems like hunger and miserable lives. Currently, it is believed that smallholder farmers can feed the world’s undernourished people [1,2]. Accordingly, agriculture employs about 62% of the population in SSA (excluding South Africa) and generates about 27% of the Gross Domestic Product [3]. Agriculture accounts for the vast majority of the poor’s livelihood activities and also holds the most promise for pro-poor economic growth [4]. Smallholder farmers, mostly women, who produce the majority of agricultural products, face various challenges, including access to adequate information, services, and key value chains [5]. Many women experience a life that is a complex web of multiple roles and multi task. This requires an average woman to conduct different roles at different times in a bid to fulfill her family needs. Women especially those in the rural areas are extensively involved in serious farm operation and agricultural activities. Women make up over half the agricultural labour force, yet they are frequently subject to discrimination. They play a vital but under-recognized and unsupported role in food production. Looking at the national average of women in the agricultural labour force, they vary but globally they have a principal role in agro business, food processing and consumer related activity [6].
In Nigeria, women consist 60 – 80 percent of labour in agriculture starting from production, processing and marketing of food. These women have their own farms and also assist in the family farms [6]. This has made the Nigerian women to be in a position to contribute to food supply. Despite outnumbering the men farmers, they are still not usually seen as the “typical farmer” by policymakers and administrators such as extension officers and are therefore often bypassed by agricultural services [7]. Women’s potential to become successful farmers have also been limited due to the fact that they are facing many other obstacles and challenges such as lack of access to productive resources, information and credit [8]. In addition, women’s social networks are often more local, connected to the rural village and therefore often more restricted than men’s, whose mobility is often greater and reaches outside the borders of the village. Since most information channels in Nigeria and Africa are built on social networks, women’s networks offer fewer opportunities for learning about, as well as engaging in, new productive opportunities and strategies, leading to underperformance of the sector, which is bad for the country and rural development at large [9]. There lies both an instrumental and intrinsic value in enabling women to optimize their production to the same extent as men [8]. When it comes to the former, research shows that women’s agricultural productivity can increase around 20-30 percent, if women would get the same kind of access to productive resources that men have.
Optimizing women’s production should be equally important based on a gender equality perspective and rights-based approach, because they should have the same right to be as productive as men, which in the end would benefit the whole country at large by reducing poverty and hunger. Applying ICT solutions could be a way to “extend the reach of existing information channels thus overcoming barriers women farmers face in accessing information” [9]. A number of studies have shown that ICT can have a positive impact on rural livelihoods and farmers, whether it is by encompassing or by accessing vital market information on a mobile phone [10,11], or by generating thematic maps and online applications to monitor the spread of agricultural pests [12]. In agriculture, information is a critical factor that has always mattered, and even though farmers may have undertaken the same activities for years, decades or even centuries, producers have not always found it easy to obtain answers when conditions for them have changed [13]. Usually for many farmers in low-income countries, information is obtained through a complex web of social networks [9]. Applying modern Information and Communication Technologies (ICTs) such as mobile phones, radios, TVs and Internet services in agriculture may offer a new way of sharing information and knowledge amongst farmers. But due to prevailing inequalities in accessing ICTs, many groups in low-income countries are often left out from using ICTs, especially women [14]. The inequality experienced by women is often referred to as the “gender digital divide”, as empirical evidence shows that women have worse access to, and use ICT less, than men, both in rich and poor countries [11].
Communication is indispensable for all endeavors to bring about a societal change. The emergence of information communication technology (ICT) has enabled to collaborate, interact and information in a fast pace which has brought a greater impact on society. The ICT is a diverse set of technological tools and resources to create, disseminate, store, bring value addition and manage information [15]. ICTs do play an important role in disseminating a wide range of information and advice leading to knowledge and attitude change among rural communities. It is also supporting rural communities to acquire new skills and is also creating new employment opportunities.
Statement of Problem
Increasing production is a major challenge facing present agriculture. Smallholder farmers which dominate the landscape of developing world need to improve farming through acquiring adequate knowledge and information [16]. Farmers exhibit ambiguous risk-averse behavior when they lack information pertaining to the likelihood of occurrence of the possible outcomes of new technology, which might have a detrimental impact on adoption (Tessema, 2016). A mix of several factors prevents the adoption of new agricultural technology and innovation by farmers and involves the level of education, individual risk preferences, capital, perception, as well as inputs such as land, labour and credit, as well as access to information [16]. Relevant and suitable information on best practices, new technologies, postharvest handling, and value-addition are key in order to boost productivity [5]. According to Asayehegn (2012), in Ethiopia most agents use individual extension methods (farm or home visits and use of contact farmers) to communicate and to disseminate agricultural technologies to farmers. Agents are also working under areas characterized by lack of infrastructural facilities such as transportation. This situation is the same in Nigeria where conventional extension methods such as farm or home visits and the use of contact farmers do not provide the needed agricultural information on timely basis (Deribe.K, 2011).
The impact of mobile phones on development will, however, in the end be determined not only by the number of owners of SIM cards and subscription rates, but also by the actual ways in which mobile phones are used and the benefits that Africans derive from using mobile phones (Bornman, 2012). Again, due to the high extension agent farmer ratio, it is practically difficult to reach the farmers by face-to-face or individual contact methods. The abovementioned problems call for the use of ICTs to support agricultural extension services, because ICTs, particularly mobile phones, can be very effective in delivering timely and relevant information to farmers, even to those living in remote areas. Though the potential of mobile phones is vast, little is known regarding the use of mobile phones in agriculture among women farmers in Nigeria.
Objectives of the Study
The main objective of this study is to analyze the roles of information communication technologies (ICTs) in improving of rural women farmers’ access to agricultural services and agribusiness in Imo state. The specific objectives of this study include to: a. identify ICT devices available to women farmers; b. identify agricultural services available to respondents in the area; determine the roles of ICTs in improving women access to agricultural services and agribusiness; and c. examine perceived constraints to respondents use of ICT devices in the area.
Methodology
The study was carried out in Orlu agricultural zone of Imo state, South-east Nigeria. Imo state is having three agricultural zones namely; Okigwe, Owerri, and Orlu (ADP, 2003). Orlu agricultural zone is made up of eleven (11) LGAs namely; Njaba, Nwangele, Orsu, Oru west, Orlu, Nkwere, Oru east, Oguta, Ideato north, Ideato South and Isu. The state lies within latitudes 4°45°N and 7°15°N and longitude 6°50°E and 7°25°E with an area of around 5,100sq/ km. It is bordered by Abia state on the East, by the River Niger and Delta state on the west, by Anambra state to the North, and River state to the south (IMSG, 2001). The estimated population of Imo state as of 2018 is 5.8million with a population density that varies from 230-1,400 people per square kilometer (NPC,2006). The rainy season spans from March to October and is bimodal with a twoweek break in rainfall in August (August break). The main annual rainfall in the state is between 25°c and 28°c with relative humidity of about 98% during the raining season and between 50% and 60% during dry season. The major arable crops grown are Cassava, Yam, Plantain, Banana, Maize, Melon, Sweet potato and Vegetables such as Okra, pepper, tomato and telfaira (ADP,2003).
Sample and Sampling Technique
Orlu agricultural zone has 10 extension blocks and 107 extension circles manned by extension agents. All ICT women user farmers in the zone constitute the population of the study. A list of all registered ICT women user farmers in the zone was obtained from ADP office in the zonal headquarters. The list has a total number of 1,100 ICT women user farmers and 10% of the total number was randomly selected which gave a total sample size of 110 ICT women user farmers.
Method of Data Collection
The study made use of both primary and secondary data. The primary data were collected from field investigation or survey using structured questionnaires and interview schedule. Secondary data sources were utilized to provide background information and other, necessary to achieve some objectives of the study. Such secondary data includes textbooks, reports, journals, publications and proceedings. Enumerators were trained and used.
Method of Data Analysis
Basically, data were analyzed using descriptive statistical tools such as mean, standard deviation, bar graph and percentages. This was used to analyze all objectives of the study. A four (4) point liker type scale of strongly agreed (SA), agreed (A), disagreed (D) and strongly disagreed (SD) assigned values of 4,3,2 and 1 was used to achieve objective 3. The scores were added together to give 10 and was divided by 4 to give a discriminating index of 2.50. Therefore, a mean of 2.50 and above was adjudged okay and accepted as role of ICTs in helping women farmers gain access to agricultural services while any value between 2.50 was not accepted. While objective (4) was analyzed on a 4 point like type scale of very important constraints (VIC), important constraints (IC), less important constraints (LIC), not important constraints (NIC) assigned scores of 4,3,2 and 1, added together to give 10 and divided by 4 to give a discriminating index of 2.50. Mean values 2.50 and above were taken as constraints while mean values below 2.5 were not constraints of ICT use in gaining access to agricultural services.
Results and Discussion
ICT devices available to women farmers
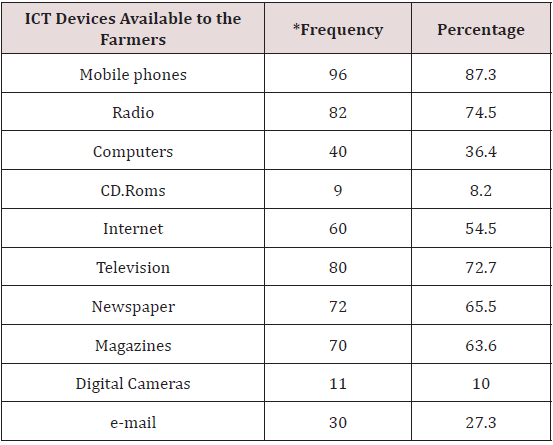
Table 1 showed the different ICT devices available in the study area. Mobile phones, radio and television are the most available ICT devices in the study area with a percentage of 87.3%, 74.5% and 72.7% respectively. This means that these devices are the most common sources of technological information and dissemination of innovation in the area. Newspapers with 65.5%, magazines and internet with percentage of 63.6% and 54.5% respectively also indicate that information can be sourced through them. Computer and E-mail with percentage of 36.4% and 27.3% respectively are also sources of information. On the other hand, digital cameras which has a percentage of 10 is not readily available to everyone. This could be due to high cost or illiteracy of the respondents. CDRoms have the least percentage of 8.2%. Not many people have this device.
Source: Field survey data 2017. *Multiple responses.
The above agrees with Singh et al (2015) reported that Agriculture Information System (AIS) is a computer-based information system which contains all the interrelated information which could really help farmers in managing information and policy decision making. The ICT devices that help facilitating farming activities encompassed applications like radio, television, cellular phones, computers, tablets and networking, hardware and software, satellite systems [17,18]. In the same way, Yimer [19]; Munyua [17] reported that radio is extensively used to inform users on agricultural topics, including new and upgraded farming techniques, production management, and market information. This shows that farmers may take advantage of using radio in the absence of technology especially rural farmers. The Internet and web-based applications are extensively used in sharing and dissemination of agricultural knowledge, marketing of goods and services.
Agricultural/agribusiness services in the study area
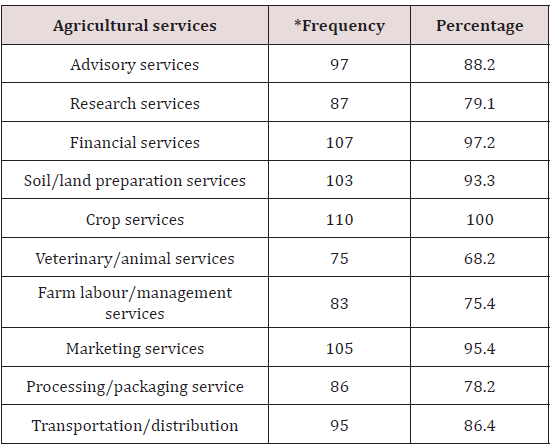
Table 2 showed that numerous agricultural/agribusiness services that exist in the study area. These services included advisory services (88.2%), research services (79.1%), financial services (97.2%), and market services (95.4%). Advisory services here means that farmers need advice on a variety of agricultural business opportunities to be explored. They need information on solving farm problems, training/education, among other services. Research services here means that farmers should be kept abreast of the latest developments/innovations in agriculture. Their problems need to be solved by research institutes which serve as centres of innovations who proffers solution and get back to them through communication with the change agent. Financial serves are very important in agric-business for farmers to optimize production and maximize income. They need credit, savings, payments/transfer and also insurance to cover crop and livestock losses. Market is important for farmers to improve their production systems, so they can fetch better prices, avoid gluts and have what it takes to grow market-led crops.
Source: Field survey data 2017. *Multiple responses.
Other services include soil/land preparation (93.3%) and crop production services (100%). Soil preparation services includes plowing, application of fertilizer, seed bed preparation and other services for improving the soil for crop planting, while crop services includes crop planting, cultivating and protecting, cultivation services, disease control of crops, entomological services, irrigation system operation systems, orchard cultivation services, seeding crops, pruning of orchard trees and vines, and weed control); crop harvesting by machine; crop preparation services for market. Animal/veterinary services (78.2%), and farm labour service (75.4%), includes animal hospitals, veterinarians and veterinary services for livestock, and animal hospitals, veterinary services for pets. Animal services, including livestock services (e.g., artificial insemination services, livestock breeding, milk testing, cattle spraying, vaccinating livestock, sheep dipping and shearing, and custom slaughtering) and animal shelters. Farm labor and management services includes farm labor contractors and crew leaders, and farm management services; and processing/ packaging services (78.2%) and transportation/ distribution services (86.4%) (e.g., sorting, grading and packing of fruits and vegetables, grain cleaning and fumigation, drying of corn, fruits and vegetables); and cotton ginning.
ICT roles in improving access to agricultural/ agribusiness
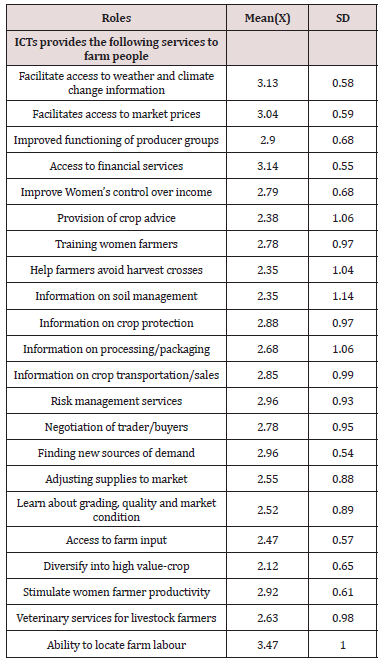
ICTs play great roles in providing access to agriculture services to women farmers. With a discriminating mean (M) index of 2.50, ICTs play the following roles - ability to locate farm labour (M = 3.47), which is usually done by making voice calls on the person, gain access to financial service (M = 3.14), which includes information on credit, loan and levies, facilitate access to weather and climate change information (M = 3.13), access to market prices (M = 3.04), risk management services(M= 2.96), finding new sources of demand (M = 2.96), improved functioning of producer groups (M = 2.90), information on crop protection (M = 2.88), information on crop transportation\sales M = 2.85, improved women’s control over income (M = 2.79). Speaking on the importance of ICTS in providing access to agricultural and agribusiness services, Chavula (2012) said ICTS help extension workers and researchers to adopt improved agricultural practices and disseminate them to farmers. They provide agricultural information that is relevant to farmers such as agricultural and farming techniques, commodity prices and weather forecasts to farmers. The utilization of ICTS especially mobile technologies help agricultural producers who are often unaware of commodity process in adjacent markets and rely on information from traders in determining when, where or for how much to sell their produce to have relevant and timely information to this regard. Delays in obtaining this information or its misinterpretation by traders has serious consequences for agricultural producers, leading to charging low prices or high/low produce supply in the markets [20]. Other areas were training women farmers (M=2.78), negotiating with trader\buyers (M=2.78), information on processing\packaging (M=2.68). Akor and Mbiti (2010) opined that ICTS facilitates information flow and can have communication between buyers and sellers leading to lower communication costs, thereby allowing individuals and firms to send and acquire information quickly and cheaper (Table 3).
Field survey, 2017 Mea score 2.50 and above accepted.
This makes market operate more efficiently, hence increase the overall production in agricultural sector and growth of the economy as a whole [21]. ICTS facilitates agricultural growth because they increase the efficiency of market interactions and provide access to real-time information mainly by enhancing farmers access to use of trading platforms over the internet through web/mobile application [22]. Finally, veterinary services for livestock farmers (M=2.63), adjusting supplies to market (M= 2.55), learn about grading, quality and market condition (M= 2.52), provision of crop advice (M= 2.38), access to farm input (M= 2.47), information on soil management (M = 2.35), help farmers avoid harvest losses (M = 2.35), diversify into high value crop (M= 2.12) and finally, stimulate women farmers productivity (M= 2.92). ). They allow people to obtain information immediately on a regular basis as compared to other information channels. The utilization of ICTS especially by using mobile phones reduces search costs, improve consumers welfare and reduces trade/market monopoly if not complete disruption of the monopolistic idea/practices.
Problems facing respondents use of ICT devices
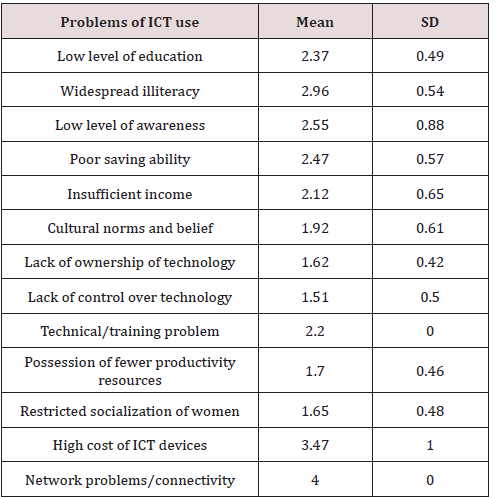
Table 4 showed that numerous problems face respondents in use of ICT devices as indicated by high mean (M) responses, which includes- network problems/connectivity (M=4.00), high cost of ICT devices (M = 3.47), widespread illiteracy (M = 2.96), low level of awareness (M =2.55), poor saving ability (M 2.47), insufficient income (M=2.12), technical/training problems of farmer part (M=2.20). This affirms FAO (2000) stand that the update and harnessing of information is limited by the lack of trained personnel or lack of access to know how. Other constraints that also faced respondents use of ICT devices included; cultural norms and belief (M = 1.92), possession of fewer productivity resources (M=1.70), restricted socialization of women (M= 1.65), lack of ownership of technology (M=1.62), lack of control over technology (M= 1.51) and low level of education (M = 1.37). The above agrees with previous studies that inadequate, and unstable power supply, cost of hardware and software are high with respect of average rural dwellers [23]. Similarly, Taragona [24] maintains that awareness, time, cost of technology, system integration, and software availability are the main constraints of ICT adoption in horticulture.
Field survey 2017. Mean response 2.50 and above accepted.
Conclusion
From the study, one can conclusively say that mobile phones, radio and television were the most accessed and utilized ICTs devices among the respondents. However, the extent of access and utilization of contemporary ICTs devices such as CD-Roms, digital camera are still very low. There was a high perception of positive effect of ICT in improving women’s access to agricultural services and agribusiness, however, the use of these ICT devices are limited by some factors. Therefore, there is need to encourage ICT user farmers in the area, by making available all that are necessary for successful ICT usage. Including training in order to promote maximum gain and utilization of technological information which will lead to improved access to agricultural services thereby improving farmers practices, increased productivity as well as bettering standard of living of the farmers.
Read More Lupine Publishers Agriculture Journal Articles: