Lupine Publishers| Journal of Biomedical Engineering and Biosciences
Abstract
In the past few years the utilization of biological signals as a method of interface with a robotic device has become increasingly more prominent. With the many of these systems being based on EEG and EMG.EMG based control has five main parts data acquisition, signal conditioning, feature extraction, classification, and control. This paper seeks to briefly cover the aspects of data acquisition and signal conditioning. After which, various methods of feature extraction, and classification are discussed.
Introduction
The use of EMG in Brain-Computer Interaction (BCI) as part of a Human-Computer Interface (HCI) is a method of control that allows for a more natural use of one's own existing muscles. Electromyography (EMG) is measured from the muscles as they receive the signal of activation from the brain. This paper presents an analysis of various methods of feature extraction and classification of the EMG signals. As EMG rapidly fluctuates with time and can contain some corruption in the data, due to noise. This means it is critical to choose the methods of feature extraction and classification to improve accuracy and to decrease the computational demand.
The methodology of EMG based control is mainly concerned with data acquisition, signal conditioning, feature extraction, classification, and then control (Figure 1) [1]. This paper presents in the next section a brief description of the method of data acquisition. Then following this will also be a brief description of signal conditioning. Next, the methods of feature extraction are presented. Each method is described with an equation and is then experimental results are presented for easy comparison. All the simulations were done in MATLAB with scripts all using the same sample size, and segment length. The following section then goes on to present different methods of classification in their formal nature. This paper then concludes with a discussion of the pros and cons of the different methods of feature extraction techniques and some specific application of those techniques. As well as a discussion of the different classifiers and some possible specific application of those classifiers.
Figure 1: Block diagram of the process of EMG processing for control.
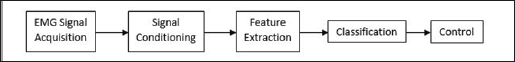
Data Acquisition
EMG data can be gathered in two different ways: invasive, and noninvasive [2]. The invasive method is performed by inserting a needle type electrode through the skin into the muscle desired. This technique is mostly used for diagnostic purposes. The invasive method provides high-resolution data, and accurate localized descriptions of muscle activity. This method, however, does cause some discomfort to the patient, and is not suited for repeated daily use. The noninvasive method uses surface mounted electrodes commonly positioned over specific muscles. This decreases the patient s discomfort and allows for the ability to be a fully portable device. The accuracy and resolution of the device depends on the sampling rate and the segment length [3]. This method has commonly used adhesives and conductive gels for the mounting of the electrodes. However, in recent years the improvement of surface mounted EMG sensors has made it possible to mount sensors without adhesive or gel.
Signal Conditioning
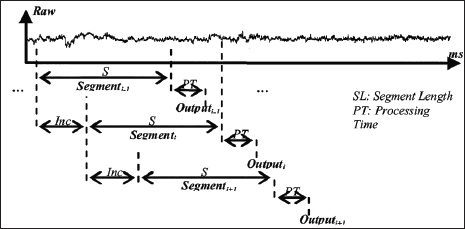
Data segmentation is done using two main methods: overlapping segmentation, and disjoint segmentation [4]. Disjoint segmentation uses separate segments with predefined length for feature extraction (Figure 2). While in overlapped segmentation, the new segment slides over the current segment, where the interval of time between two consecutive segments is less than the segment length and more than the processing time (Figure 3). This shows that disjoint segmentation of data is associated with segment length. While overlapped segmentation of data is associated with segment length and increment [5]. In experiments done by Oskoei, and Hu [4], disjoint and overlapped segmentation was compared to display their classification performance. The length of 50ms was used in disjoint segments whereas overlapped systems used segments having a length of 200ms with an increment of 50ms. The results showed that the defined disjoint segmentation 200ms provided high performance in EMG classification and an adequate response time allowing for real-time use. With the defined overlapped segmentation shortening the response time without noticeably degrading the accuracy of data. This indicates that to maintain an efficient use of computational resources while not compromising the accuracy of data, it is imperative to implement an appropriately timed method of overlapped segmentation.
Figure 2: Graphical representation of disjoint segmentation [4].
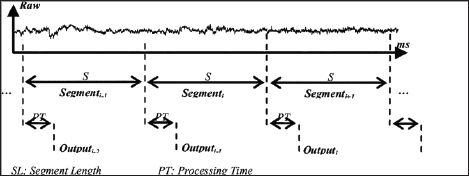
Figure 3: Graphical representation of overlapping segmentation [4].
Feature Extraction
Feature extraction is the transformation of the raw signal data into a relevant data structure by removing noise, and highlighting the important data. There are three main categories of features important for the operation of an EMG based control system. Those being the time domain, frequency domain, and the time-frequency domain [1,5]. However, due to the intense computation needs of transformations required by the features in the time-frequency domain, this method is not used for therapeutic devices.
Time Domain
Features in the time domain are more commonly used for EMG pattern recognition. This is because they are easy, and quick to calculate as they do not require any transformation. Time domain features are computed based upon the input signals amplitude. The resultant values give a measure of the waveform amplitude, frequency, and duration with some limitations [6]. The methods of integrated EMG, mean absolute value, mean absolute value slope, Simple Square integral, variance of EMG, root mean square, and waveform length will be discussed in more detail in the following sub-sections. Each consecutive section will reuse the same notation for better understanding. For each method, a simple test was done with MATLAB scripts for sake of comparison.
Read More About Lupine Publishers Journal of Biomedical Engineering and Biosciences Please Click on Below Link: https://biomedical-sciences-lupine-publishers.blogspot.com/
No comments:
Post a Comment
Note: only a member of this blog may post a comment.