Lupine Publishers| Journal of Drug Designing & Intellectual Properties
Abstract
Serotonin, 5-hydroxytryptamine, represents a class of monoamine neurontransmitters, all of which having a chemical template comprised of a basic amino group separated from an aromatic nucleus by a two-carbon aliphatic chain. In this paper we present a docking study on serotonin targeting the proteins 3ADX and 2YX8, respectively, performed by AutoDock Vina. A set of thirtyfive serotonins, downloaded from PubChem, was modeled, within the hypermolecule strategy; the predicted activity was LD50 and prediction was done on similarity clusters with the leaders chosen as the best docked ligands on the Peroxisome proliferatoractivated receptor gamma. It was concluded that LD50 of the studied serotonins is not directly influenced by their binding energies to the target proteins.
Keywords: Serotonin; Docking; Binding affinity; 3ADX; 2YX8; Hypermolecule; LD50; QSAR
Introduction
Serotonin is a neurotransmitter involved in a wide variety of behaviors, including feeding and body-weight regulation, social hierarchies, aggression and suicidality, obsessive compulsive disorder, alcoholism, anxiety, and affective disorders [1-3]. Peroxisome proliferator-activated receptors are ligandactivated transcription factors that regulate genes important in cell differentiation and various metabolic processes, especially lipid and glucose homeostasis [4].
Receptor activity-modifying proteins (RAMPs) are a class of proteins that interact with and modulate the activities of several Class B G Protein-Coupled Receptors including the receptors for secretin, calcitonin (CT) and glucagon, RAMPs complex with Sumatriptan molecule [5].
A method is described to dock a serotonin into a binding site in a peroxisome proliferator-activated receptor gamma on the basis of the complementarity of the inter-molecular atomic contacts. Docking is performed by maximization of a complementarity function that is dependent on atomic contact surface area and the chemical properties of the contacting atoms [6].
Serotonin is metabolized in cells of the brain, gastrointestinal tract, liver, lungs and platelets; it exerts its effect on endothelial cells through multiple receptors [7].
A pharmacophore model can be established either in a ligand based manner, by superposing a set of active molecules with low binding energy [8] and extracting common chemical features that are essential for their bioactivity, or in a structure-based manner, by probing possible interaction points between the protein target and serotonins [9,10].
In our previous work [11], we performed a QSAR study on a set of serotonins, by the similarity cluster prediction approach, proposed by TOPO Group Cluj. In this paper, a docking study, performed to identify the geometric description of the pharmacophore in the interaction of this class of ligands with the peroxisome proliferatoractivated receptor gamma, is reported. We developed a new approach by creating similarity clusters using as leaders those ligands showing the best scores in the docking test.
Docking Study
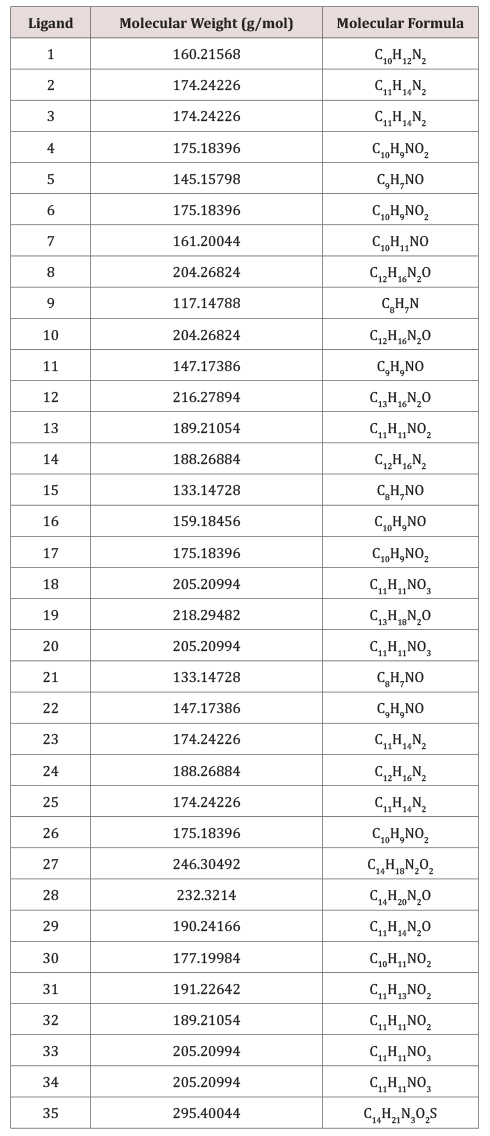
A set of 35 derivatives of serotonin were taken from PubChem Database [12] and were divided into a training set (25 molecules) and a test set (10 molecules, in Italics), taken with the lowest docking energy (Table 1). The property chosen for modeling was LD50 (on rat, intraperitoneal route administered).
The molecular docking study was carried out to explore the binding mode of serotonin derivatives (Table 2) within the binding energy of peroxisome proliferator-activated receptor gamma 3ADX and Receptor activity-modifying protein 1 2YX8.
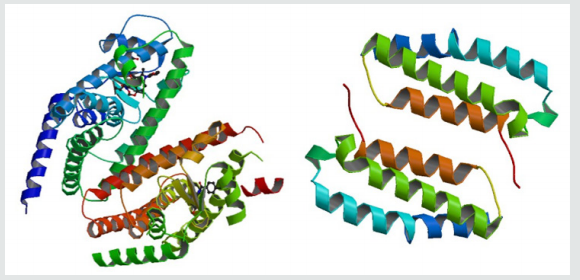
Figure 1:The proteins (Peroxisome proliferator-activated receptor gamma left, Receptor activity-modifying protein 1 right) (RCSB PDB CODE: 3ADX, 2YX8).
Molecular graphics laboratory (MGL) tools and AutoDock4.2 was downloaded from www.scripps.edu [14]. We employed the Lamarckian genetic algorithm (LGA) for ligand conformational searching [15]. AutoDock Tools was used for creating PDBQT files from traditional PDB files. The grid menu is toggled [16]; after loading protein. pdbqt the map files were selected directly with setting up the grid points appropriate for the searching of ligand within the active site of the protein molecule. This way the grid parameter files are created with setting up the map files directly. The docking parameter files were completed by using the Lamarckian genetic algorithm [17].
(Figure 2) illustrates the final Lamarckian genetic algorithm docked state: binding energy of ligands with the active site of Peroxisome proliferator-activated receptor gamma (3ADX). Docking energies lie in the range: -7.0 and -5.1kcal/mol. It seems that the higher the number of rotatable bonds, the higher the ligand–enzyme affinity is.
The distribution of the docking energy (Figure 2) may be attributed to the differences in position of the functional groups within the studied compounds. Interaction of the ligand with the protein can be seen in (Figure 3).
Read More Lupine Publishers Journal of Drug Designing & Intellectual Properties Please Click on Below Link: https://lupine-publishers-drug-designing.blogspot.com/
https://fashion-technology-lupine-publishers.blogspot.com/


No comments:
Post a Comment
Note: only a member of this blog may post a comment.