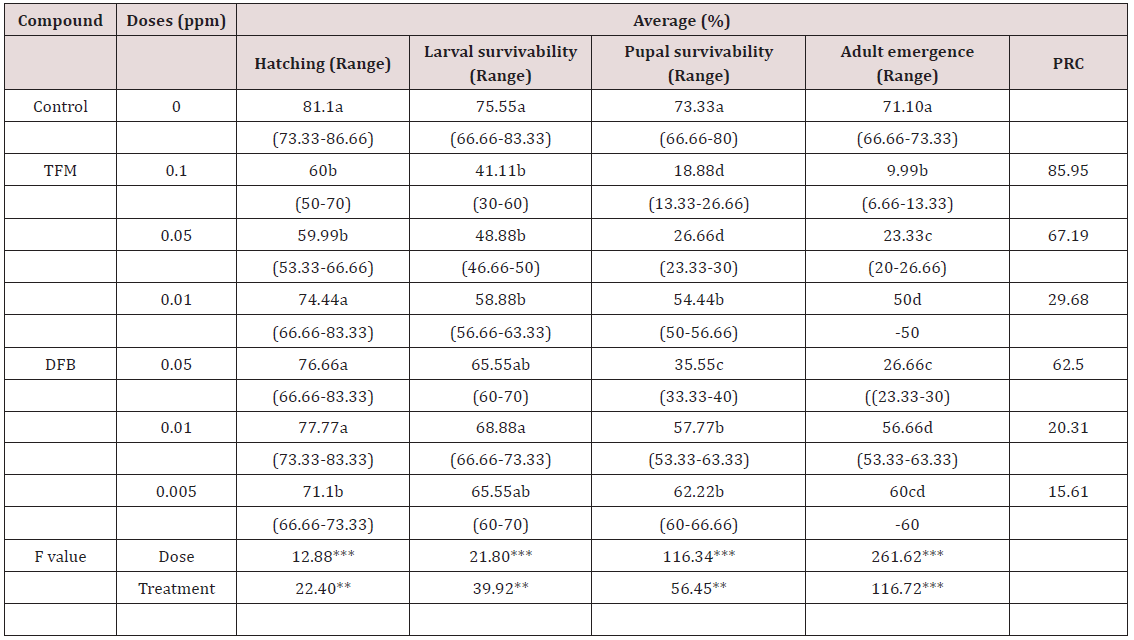
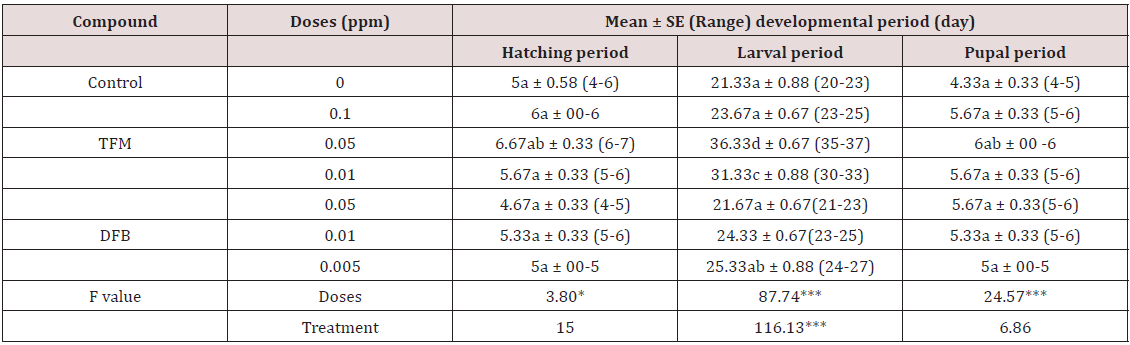
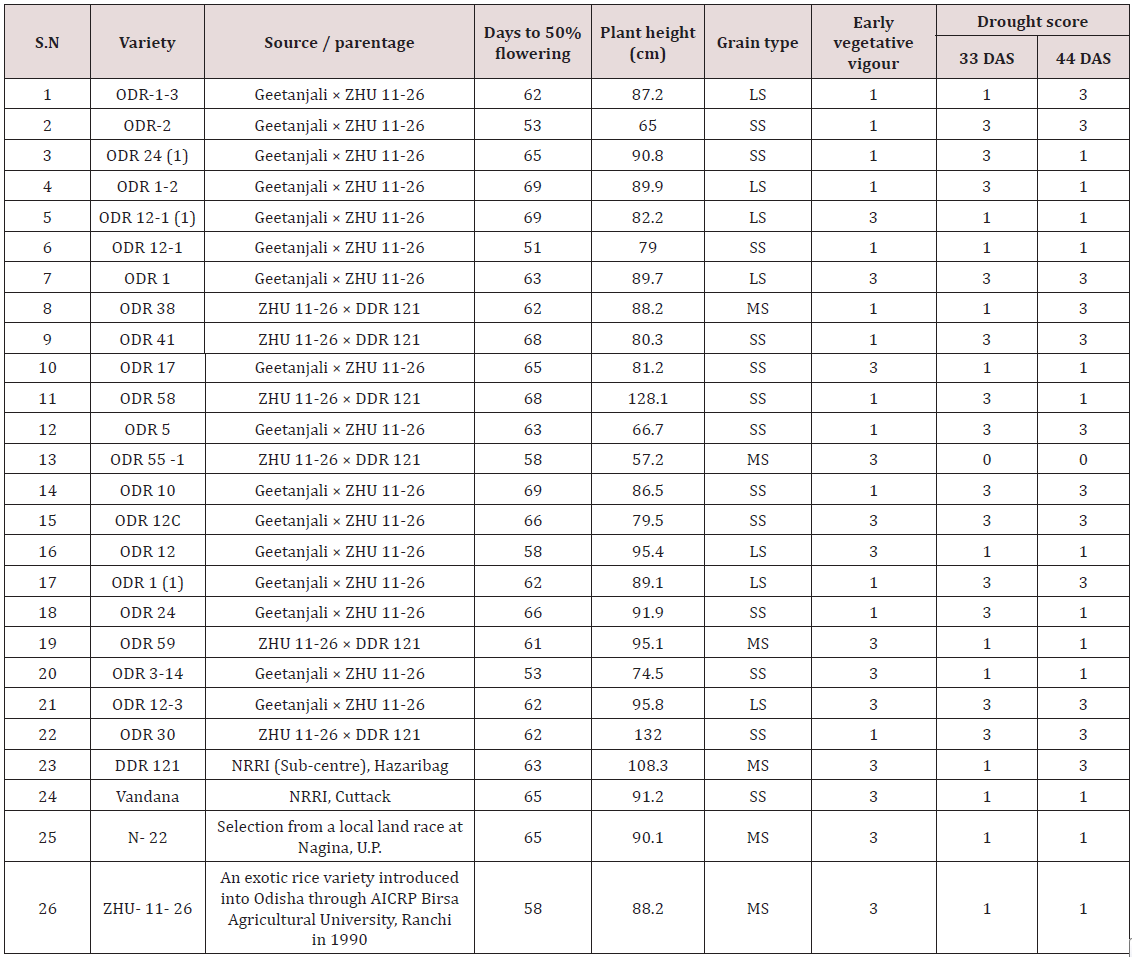
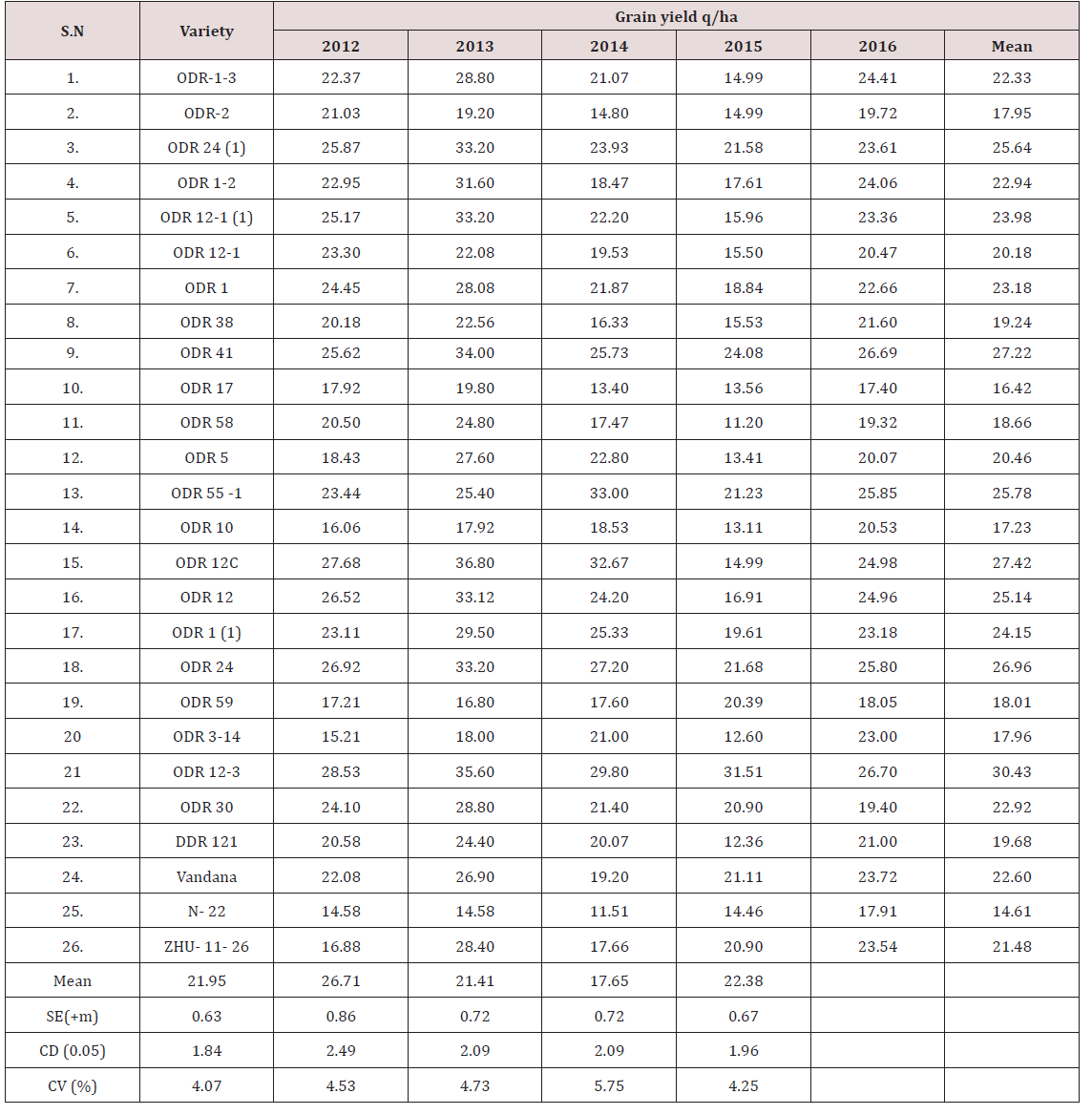
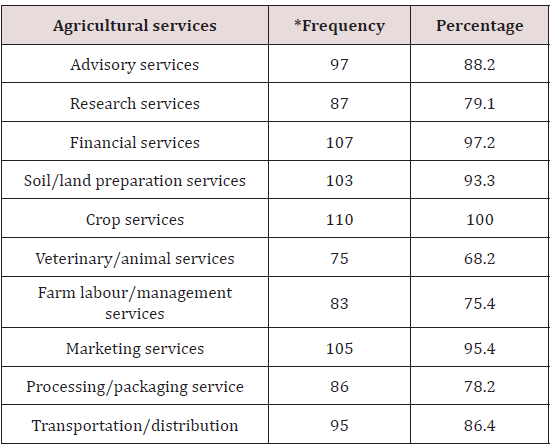
Lupine Publishers | Current Investigations in Agriculture and Current Research
Abstract
The present study was an initial attempt towards enhancing grain
yield in rice through molecular breeding approach by allele
mining. Forty one diverse rice genotypes were used for allele mining
study with an objective to detect among them superior alleles
for rice grain dimensions viz. grain length, grain width, grain
thickness, grain size and grain weight. SSR markers associated with
reported genes for these traits were used for PCR amplification. It was
found that SSR primer RDD 1-2 can be used for identifying
alleles enhancing grain length from diverse genomic sources for crop
improvement. The allele A (600 bp) from Rdd 1 can be used
in breeding programmes to enhance grain length in rice. The genotypes
viz. Hans raj, Pant dhan-18, Pant dhan-10, Narendra, Pant
dhan-12, Pant dhan-4, ARR-09, Shahsarang, IVT-ASG-2712, IVT-ASG-2701,
AVT-I-ASG-2602, AVT-I-ASG-2609 and IVT-ASG-2705
identified carrying this allele can be used as a source for large grain
length in future. The primers RM 478 for grain thickness and RM
574-2 for grain width can be used in marker assisted breeding
programmes. Allele A (205 bp) of RM 478 and allele B (240 bp) of RM
574-2 associated with low grain weight and grain width, respectively can
be used for rejection at seedling stage in marker assisted
breeding programme. This study also identified novel alleles (alleles
distinct from those reported in biparental programme) which
could be significant in a larger panel of rice genotypes.
Keywords: Allele mining; rice;Oryza sativa; O. nivara; SSR primers
Introduction
A panel of 41 rice genotypes consisting of the released cultivars
of O. sativa, landraces and nineteen accessions from AVT, AVIT and
IVT series and two accessions RWR-19 and RWR-125 from wild
species O. nivara were used for allele mining study for grain traits
contributing to grain yield. The mean data for grain length, grain
width and 1000 grain weight was taken for all the genotypes used
for allele mining study. The 41 rice genotypes were categorized
into three groups in order of their grain dimensions and 1000 grain
weight. The genotypes consisted of 39.02 % large (2.49-3.62), 51.22
% medium (2.01-2.44) and 9.76 % small (1.85-1.97) for grain width
in mm; 19.51 % large (9.18-10.31), 56.1 % medium (7.36-8.74) and
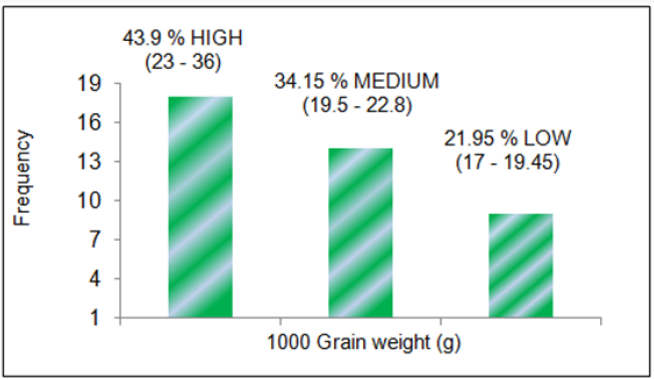
24.4 % small (5.48-6.82) for grain length in mm; 43.9 % high (23-
36), 34.15 % medium (19.5-22.8) and 21.95 % small (17-19.45) for
1000 grain weight in grams. F-test was conducted for analysis of
variance in phenotypic traits between the genotypes. Significant
variation at 5% level was observed for grain length (F=9.61) and
grain width (F=6.10). Allele mining for grain yield contributing
traits viz. grain length, grain weight and grain width was done for
the reported gene Rdd1 and the QTLs tightly associated with the
traits with an aim to identify alleles of genes conferring the grain
yield traits. The genes/QTLs associated with these three-grain
yield contributing traits viz. grain length, grain width and 1000
grain weight targeted in this study included Rdd 1 (Rice dof daily
fluctuations 1), gw8 (Grain width 8), gt7 (Grain thickness 7), gw
8.1 (grain weight 8.1), G S5 (Grain size 5), and gw7 (Grain weight
7), all previously reported to be associated with grain yield traits in
biparental studies (except Rdd 1).
Scoring of PCR products was done through gel electrophoresis
by comparing the size of the bands of the PCR products using a
gel documentation system. The markers showing bands of similar
positions in gels for all the genotypes were monomorphic markers
while those markers showing bands at different position were
polymorphic markers. All the genotypes that show products of
similar size with that of expected amplicon size for the reported
trait will carry the desired allele for grain yield and likewise the
alleles for all the target regions were identified. PCR-based allele
mining for these traits was performed in a panel of 41 genotypes
using 10 microsatellite markers reported to be tagged with these
genes/QTLs. Ten microsatellite markers were used for allele mining;
out of which nine markers (RDD1-2, RM 234, RM 478, RM 23201,
RM 502, RM 574-1, RM 574-2, RM 593-1 and RM 593-2) amplified.
Out of these nine markers, RM 23201 was monomorphic and the
rest were polymorphic. Maximum number of four alleles was found
for marker RM 478 and RM 574-2, three alleles for RDD I-2 and
RM 234 and two alleles for RM 502, RM 574-1, RM 593-1 and RM
593-2 when scored in agarose gel. Chi square test was conducted
to study the association of the markers with grain traits [1]. Each
of the alleles observed in the top 15 and bottom 15 genotypes
were tested for the consistency of their distribution with the whole
population. Significant associations were observed for allele A in 15
largest genotypes (Χ2=3.94, p=0.05) and B allele (Χ2=4.51, p=0.03)
in the 15 smallest length genotypes for primer RDD 1-2. Allele A
was found to be associated with large grain length and allele B was
found to be associated with small grain length. The genotypes viz.
Hans raj, Pant dhan-18, Pant dhan-10, Narendra, Pant dhan-12,
Pant dhan-4, ARR-09, Shahsarang, IVT-ASG-2712, IVT-ASG-2701,
AVT-IASG-2602, AVT-IASG-2609 and IVT-ASG-2705 were found to
contain A allele found to be associated with large grain length for
RDD 1-2 marker. Significant association was observed in marker
RM 478 with grain weight and for distribution of A allele in the
bottom 15 genotypes (lowest grain weight) with the distribution in
the overall population (Χ2=5.08, p=0.02) and for primer RM 574-2,
distribution of B allele (Χ2=4.3, p=0.08) in the bottom 15 genotypes
(smallest grain width) was observed to be significant.
From this present experiment, hybrid plants from inter-specific
crosses can be used for backcrossing in future plant breeding
programmes. Crosses can also be attempted for genotypes that
failed during this experiment. Primer RDD 1-2 can be used for
identifying alleles enhancing grain length from diverse genomic
sources for crop improvement [2]. The allele A (600 bp) from Rdd
1 can be used in breeding programmes to enhance grain length
in rice. The genotypes viz. Hans raj, Pant dhan-18, Pant dhan-10,
Narendra, Pant dhan-12, Pant dhan-4, ARR-09, Shahsarang, IVTASG-
2712, IVT-ASG-2701, AVT-IASG-2602, AVT-IASG-2609 and
IVT-ASG-2705 were identified to be carrying this allele can be used
as a source for large grain length in future. The primers RM 478
for grain thickness and RM 574-2 for grain width can be used in
marker assisted breeding programmes. Allele A (205 bp) of RM
478 and allele B (240bp) of RM 574-2 associated with low grain
weight and grain width, respectively can be used for rejection at
seedling stage in marker assisted breeding programme. This study
also identified novel alleles (alleles distinct from those reported in
biparental programme), which could be significant in a larger panel
of rice [3].
Materials and Methods
A panel of different rice genotypes including released cultivars
of O. sativa i.e., IR-1552 (LR-55), Govind, CAU R-1, Kalanamak,
Hansraj, Pant dhan-4, Pant dhan-10, Pant dhan-12, Pant dhan-16,
Pant dhan-18, Narendra-359, landraces like ARR-09, LR-1 (Balwai),
LR-5 (Laljagli), LR-23 (Sabhagidhan), LR-71 (Basmati paddy), LR-
77 (BR-1),Shahsarang, Bhasphool (LR-2), Chakaoporieton (LR-
26), and nineteen accessions from AVT and IVT series and two
accessions RWR-19 and RWR-125 from wild species O. nivara were
used for allele mining. The experimental material seed of Pant dhan
was obtained from Pantnagar and seed of AVT, AVIT and IVT series
have been procured from DRR (Directorate of Rice Research),
Hyderabad (Tables 1-3) [4].
Table 1: Description of different rice (O. sativa and O. nivara)
genotypes used for allele mining.
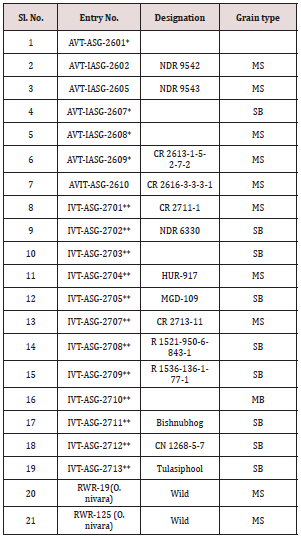
*- Advanced varietal trial; **- Initial varietal trial; SB: Short bold;
MS: Medium slender/Mild scented; LS: Long slender; SS: Strong
scented.
Table 2:
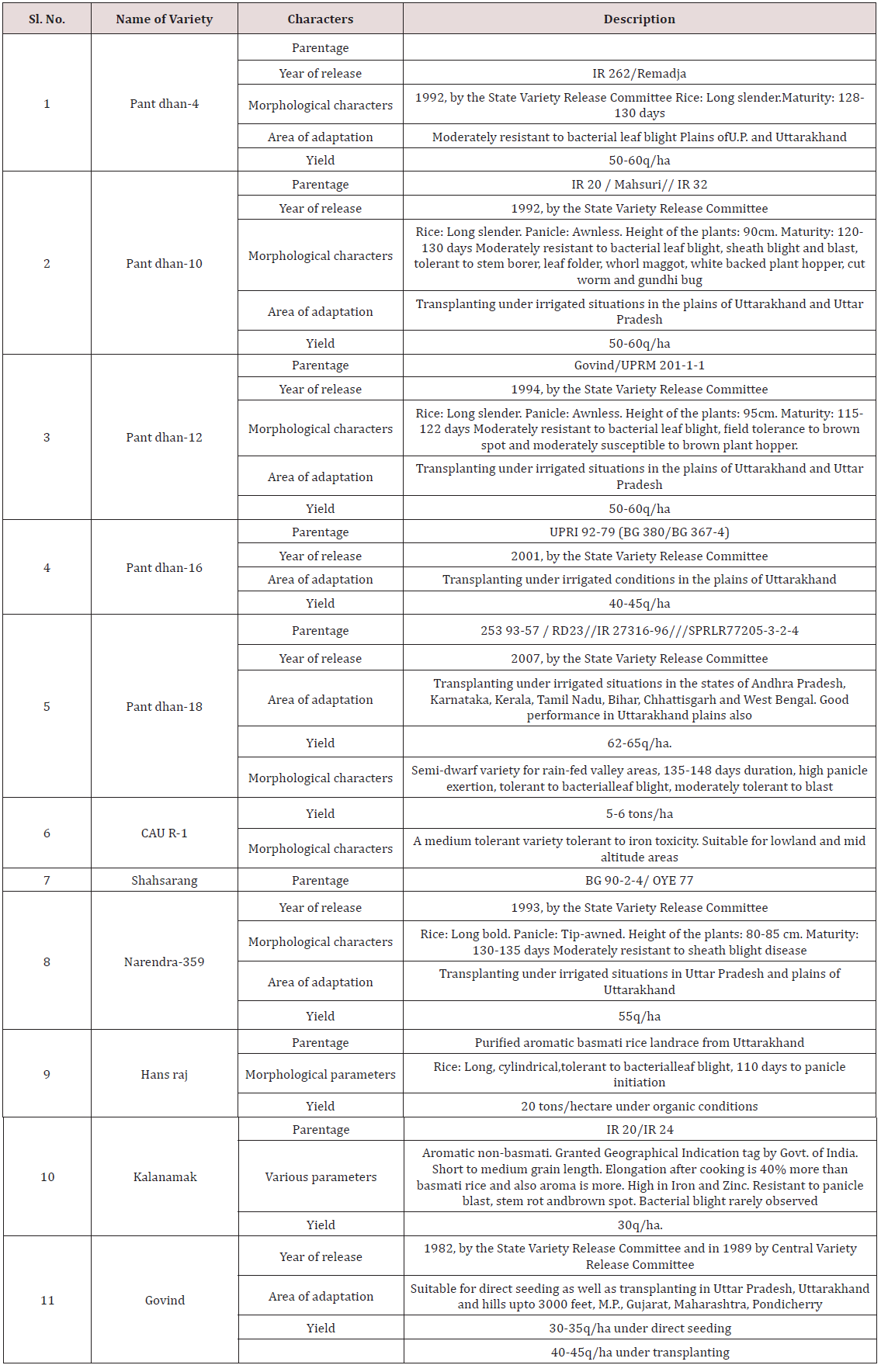
Table 3:
Statistical analysis of seed parameters for grain size
variance in 41 rice genotypes
The grain size of rice is contributed collectively by a complex
of yield related traits of which grain length, grain width and grain
weight also play a major role. Given prior concern to these three
traits, a set of data for average grain length, grain width and grain
weight has been taken for all the genotypes used for allele mining.
Phenotypic analysis/Analysis of variance
The frequency distribution in percent for grain length, grain
width and grain weight was determined for large, medium and
small grains in the population of 41 genotypes. Analysis of variance
for significance of phenotypic variations for grain length and grain
width of 41 genotypes was studied through F-test (Table 4).
Table 4:
Germination and sowing of seeds
Sowing of seeds used for allele mining study was done in the
month of February 2014 (Table 4). Prior to sowing, seeds of all the
genotypes were first allowed to germinate in sterilized Petri plates
containing filter papers moistened with water. Upon germination,
the seeds were transferred to shallow plastic trays and the
genotypes were grown in rows with each genotype in a single row
having at least twenty seedlings per row [5]. The soil was finely
prepared free of clods and was mixed with farm yard manure. The
trays containing the plants were kept inside the greenhouse and
the plants were maintained by regular watering and also weeding
as and when required.
Molecular Characterization
SSR analysis for polymorphism
Rice dof daily fluctuations1 (Rdd1), Grain width5 (GS5), Grain
thickness (gt7) and Grain weight8 (GW8) previously reported
to be linked to the grain traits/QTLs were used for allele mining
studies in different rice genotypes. Forty one rice genotypes
including two accessions of O. nivara were used for allele mining
studies to detect the existing alleles among the genotypes for the
given traits. Markers for these reported genes were obtained from
Gramene marker database. Genomic DNA from rice leaves of all
the genotypes were isolated using CTAB method of Doyle [3]. The
DNA samples were diluted to obtain a working standard according
to the concentration and the quality of the DNA obtained. The PCR
reaction conditions for primers were standardized to obtain good
quality bands and were used as the standard reaction conditions
throughout the whole PCR amplification process [6].
The three yield contributing traits namely grain length, grain
width and grain weight was targeted in this study. The gene Rdd1
has been reported to be indirectly related to increase in grain size
in rice. Since increase in grain size is constitutively contributed by
grain traits which include grain length, the RDD1 markers associated
with this gene has been used in allele mining for grain length trait.
Gs 5 is a reported QTL which has been known to associate positively
with grain width and thus contributing to grain size and the RM
markers reported to be associated with this gene and flanking at
the regions near it were used for allele mining study. Another QTL,
gw 8, known to positively control grain width and QTLs; gt7, gw
8.1 and gw 7 for grain weight were targeted for allele mining in the
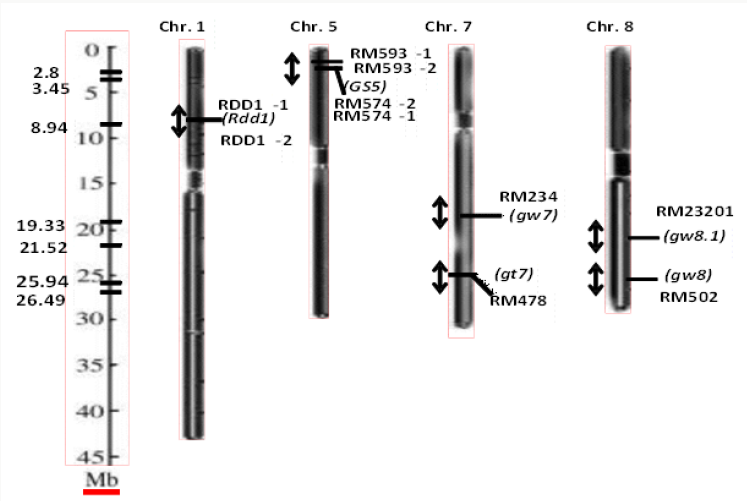
present study (Figure 1).
Figure 1: Chromosome map showing genes/QTLs (italicized) associated with grain yield traits and the position of the markers
(designated as RDD 1 and RM) linked to these traits respectively, in rice. The scale of the maps in megabase is shown on the
left axis.
Isolation of Genomic Dna from Plant Tissue
Genomic DNA was isolated from genotypes used for allele
mining. Tender leaves from young rice seedlings were used for
the extraction of genomic DNA. Genomic DNA from rice leaves was
isolated using CTAB method of Doyle [3]. Young actively growing
leaves of 15 days-old plants were collected and used for DNA
extraction.
Procedure for DNA extraction
About 2g of young fresh plant leaf tissue was rapidly frozen
in liquid nitrogen and ground to a fine powder in a pre-chilled
sterile pestle and mortar. The frozen powder was transferred to a
2ml tube. 1ml of CTAB extraction buffer was added to each tube
and tubes were inverted several times with gentle shaking. The
tubes were incubated at 65 0C in a water bath for 1 hr. Contents
were mixed by inverting the tubes after every 10-15 minutes. The
tubes were cooled to room temperature and 1 ml of chloroform:
isoamyl alcohol in the ratio of 24:1 was added to each. The contents
were mixed by inverting the tubes several times. The tubes were
centrifuged at 10,000 rpm for 10 minutes at 4 0C. The upper clear
aqueous supernatant was removed and transferred to fresh tubes.
1ml of pre-chilled isopropanol was added to each tube and the
contents were mixed by inverting the tubes several times and the
tubes were kept overnight at -20 0C. Although DNA precipitation
began as soon as isopropanol was added, it was kept overnight for
better and complete precipitation. The samples were centrifuged
for 10 minutes at 5,000 rpm at 4 0C. The solution was poured
without disturbing the DNA pellet at the bottom of the tubes. 1.5ml
of 70% ethanol was added and then centrifuged at 5,000rpm for 5
minutes at 4 0C. Supernatant was decanted. Pellet was air dried for
15-20 minutes. 50μl of TE buffer was added to the pellet [7].
DNA quantification
The quantification of DNA was done by staining DNA with
ethidium bromide after electrophoresis in 0.8% agarose gel at
100V for 1 hour in TBE buffer (0.04M Tris borate, 0.001 M EDTA,
pH 8.0) using known DNA concentration standard.
Dilution of DNA samples
A part of the DNA sample of each genotype was taken and
dilution of each sample was made with appropriate amount of TE
buffer to yield a working concentration and stored at -20°C for
further use in PCR amplification.
Polymerase chain reaction (PCR)
Dilution of SSR markers: The SSR markers which were in
lyophilized state were centrifuged and diluted with TE buffer
accordingly to yield 100pmol/μl stock concentration and from the
stock, working concentration of 10pmol/μl was made by diluting at
1:9 ratio of stock with TE buffer. The primer stock was immediately
transferred to -20 °C to prevent degradation during longer duration
storage and the working standard was kept in 4 °C for use in the
study.
Standardization of PCR conditions
Standardization of annealing temperature: Annealing
temperatures for each of the primers was standardized by PCR
reaction at least three different PCR conditions. The reaction
condition giving amplification with good bands were taken as the
standard for all the following experiments.
PCR conditions for markers used for allele mining
Primary denaturation: 95 °C for 3 minutes
Denaturation: 95 °C for 0.30 minutes
Annealing: 55 °C for 0.45 minutes
Extension: 72 °C for 0.40 minutes
Final extension: 72 °C for 2 minutes
Number of cycle: 32
Allele mining of reported genes/ QTL for yield related
grain traits in rice using SSR markers
In this study, 41 diverse rice genotypes were used for allele
mining study with an objective to detect among them superior
alleles for rice grain dimensions viz. grain length, grain width, grain
thickness, grain size and grain weight. SSR markers associated with
reported genes for these traits were used for PCR amplification
Micro satellite markers linked to reported genes/QTL for grain
yield traits were used to detect polymorphism and to detect the
yield enhancing alleles among 41 rice genotypes. PCR amplification
was done with a set of 10 primers (Tables 5 & 6) and analysis for
polymorphism was done through electrophoresis in 3% agarose
gel. Scoring of PCR products was done through gel electrophoresis
by comparing the size of the bands of the PCR products using a
gel documentation system. The markers showing bands of similar
positions in gels for all the genotypes were monomorphic markers
while those markers showing bands at different position were
polymorphic markers. All the genotypes that show products of
similar size with that of expected amplicon size for the reported
trait will carry the desired allele for grain yield and likewise the
alleles for the entire target regions were identified [8].
Table 5: Concentrations of reaction mixtures for markers used for allele mining.
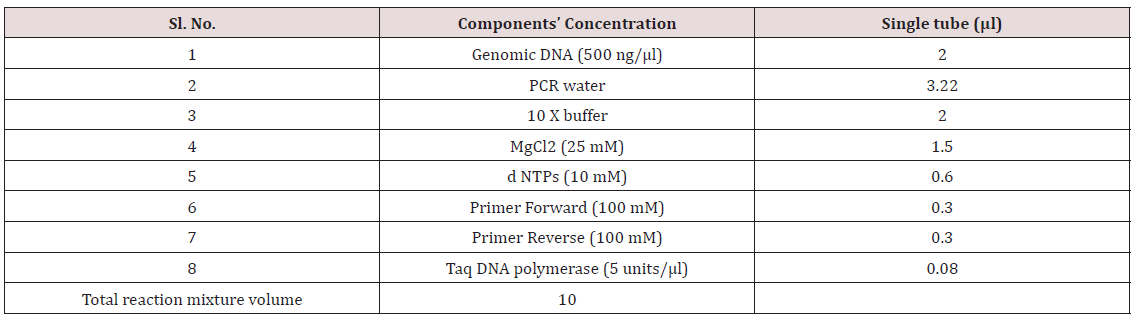
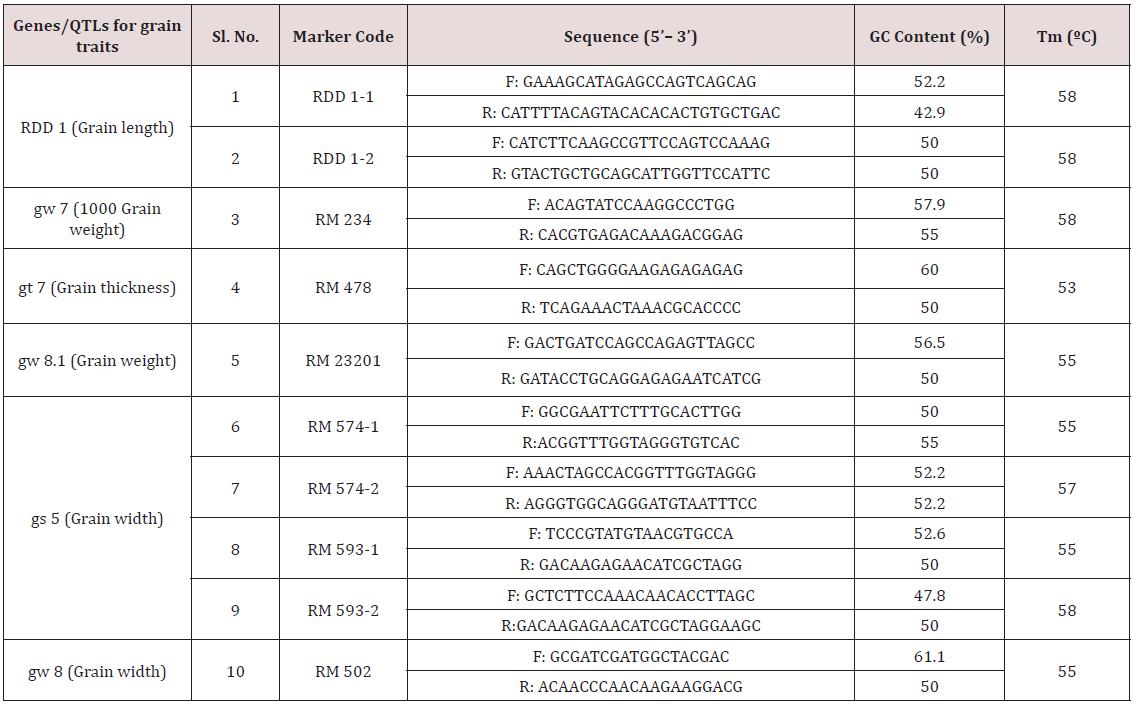
Table 6: List of SSR primers used for allele mining study.
Association between phenotype and genotype
The average grain length, grain width and 1000 grain weight
distribution in the genotypes carrying different alleles was
determined by checking whether the error bars referring to
confidence interval showed overlapping or not. The 41 genotypes
were further categorized into top and bottom groups of 15
genotypes based on grain length, grain width and grain weight. Chi
square analysis was performed to study the association between
markers with grain traits and allelic association with the trait for
each of the sub-groups.
Results and Discussion
Phenotypic analysis for yield related traits
Phenotypic variation was observed for grain dimensions in
all the genotypes. The mean data for grain width (Figure 2), grain
length (Figure 3) and 1000 grain weight (Figure 4) was taken for all
the 41 genotypes used for allele mining study. The genotypes with
larger grain width (mm) were LR-1 (3.62), LR-71 (3.07), Pant dhan-
16 (2.85), IVT-2712 (2.79) and RWR-19 (2.77). Smaller grain width
(mm) was observed in Kalanamak (2.01), AVT-2607 (2.0), Hans raj
(1.97), AVT-2701 (1.94), IVT-2703 (1.904) and AVT-2609 (1.85).
The panel consisted of 39.02 % large (2.49-3.62), 51.22 % medium
(2.01-2.44) and 9.76 % small (1.85-1.97) for grain width (in mm)
trait. The genotypes with larger grain length (mm) were Hans
raj (10.31), Pant dhan-18 (9.83), Pant dhan-10 (9.7), Narendra
(9.63), Pant dhan-12 (9.53), RWR-125 (9.27), Pant dhan-4 (9.23)
and Govind (9.12). Smaller grain length (mm) was observed for
genotypes IVT-ASG-2708 (6.41), Kalanamak (6.35), IVT-ASG-2711
(6.15), IVT-ASG-2607 (5.92), IVT-ASG-2713 (5.71), IVT-ASG-2703
(5.69), LR-71 (5.59) and LR-77 (5.48). The genotype panel consisted
of 19.51 % large (9.18-10.31), 56.1 % medium (7.36-8.74) and
24.4 % small (5.48-6.82) for grain length (in mm).The genotypes
having the highest grain weight (g) were LR-1 (36.04), Pant dhan-
18 (27.97), Pant dhan-4 (26.39), LR-23 (25.32), RWR-125 (25.05),
Narendra (24.2), Pant dhan -12 (23.97) and Pant dhan-16 (23.39).
Lower grain weight (g) was observed for genotypes, Kalanamak
(12.17), LR-77 (12.14), AVT-IASG-2609 (12.00), IVT-ASG-2713
(11.96), AVT-IASG-2607 (10.74) and IVT-ASG-2703 (10.72). The
genotype panel consisted of 43.9 % high (23-36), 34.15 % medium
(19.5-22.8) and 21.95 % small (17-19.45) for 1000 grain weight
(g). The phenotypic variance observed for the 41 rice genotypes
were found to be significant at 5% level for grain width (F=9.61)
and grain length (F=6.1).
Figure 2: Classification of rice genotypes in large, medium and small grain width categories.
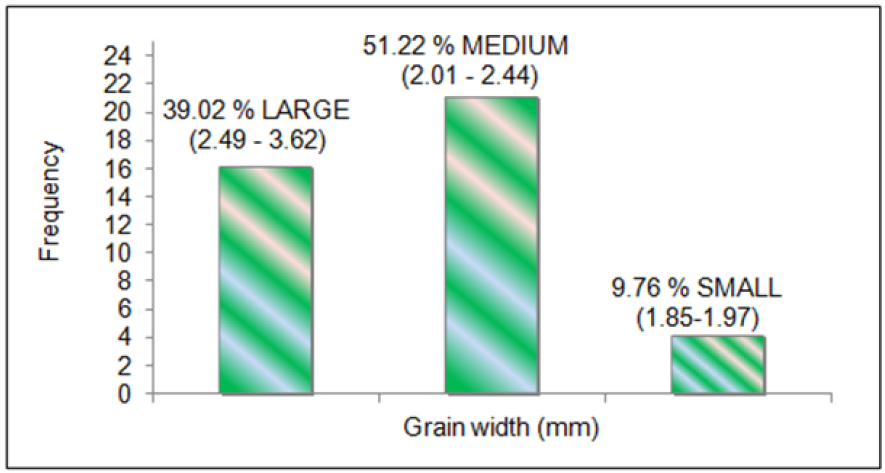
Figure 3: Classification of rice genotypes under large, medium and small grain length categories.
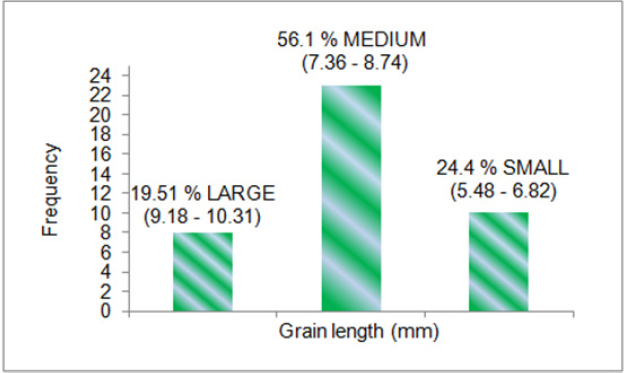
Figure 4: 4: Classification of rice genotypes in high, medium and low grain weight categories.
Allele mining for genes/QTLs
Alleles are the alternative DNA sequences at the same physical
locus which may or may not result in different phenotypic traits.
Increasing demand of rice varieties with higher yield is making
scientists work for the identification of superior and novel alleles
to be used in breeding programmes. Allele mining is another
very important molecular approach towards identifying and
bringing together useful traits from diverse genetic sources
and introgression these useful alleles into a common recipient
promising cultivar lacking one or two superior alleles for some
agronomic traits. Molecular markers are effectively being used to
detect the variant alleles present in diverse genotypes or cultivars
which are phenotypically different from each other for the traits
in question. Microsatellite markers or SSR markers are PCR-based
markers that specifically bind to the complementary sequence of
the plant genomic DNA during PCR reaction. Through allele mining
novel genes could be identified from diverse sources and used in
improvement of crops for specific traits. Allele mining for grain
yield related traits in large number of genotypes/ cultivars would
help in identifying new alleles of the genes known for the target
trait and their sources. It can also provide an insight into molecular
basis of novel trait variations and identify the nucleotide sequence
changes associated with superior alleles [9].
The quantitative trait locus (QTL) mapping has contributed to
a better understanding of the genetic basis of many agronomically
important traits such as grain yield. Grain yield is a quantitatively
inherited trait which is the result collectively contributed by several
individual grain traits. Of these, grain length, grain width and grain
weight are some of the important yield contributing grain traits in
rice. Today, a number of genes/QTLs associated with these traits
have been identified, sequenced, mapped and many associated
markers to these QTLs have been developed recently. In this study,
a set of 41 diverse genotypes including two accessions viz. RWR-
19 and RWR-125 from wild species O. nivara were used for allele
mining using nine SSR markers tagged to reported genes/QTLs.
Out of the nine primers viz. RDD 1-2, RM 234, RM 478, RM 23201,
RM 502, RM 574-1, RM 574-2, RM 593-1 and RM 593-2 used in the
present study, RM 23201 was found to be monomorphic and the
rest eight primers were polymorphic.
Association between genotype and phenotype
Chi square test was conducted to study the association of the
markers with grain traits. Each of the alleles observed in the top 15
and bottom 15 genotypes were tested for the consistency of their
distribution with the whole population. Significant association
(Χ2=3.94, p=0.05) was observed for allele A in the 15 genotypes
having large grain length for primer RDD1-2. Allele Bfor Rdd1
gene too showed significant association (Χ2=4.51, p=0.03). Allele
A was found to be associated with large grain length and allele
B was found to be associated with small grain length. Significant
association was observed for marker RM 478 with grain weight and
for the distribution of A allele in the bottom 15 genotypes (lowest
grain weight) with the distribution in the overall population
(Χ2=5.08, p=0.02) and for primer RM574-2, distribution of B allele
(Χ2=4.3, p=0.08) in the bottom 15 genotypes (smallest grain width)
was found to be significant.
Rdd 1 (Rice dof daily fluctuations1) Dof gene in rice has
been reported to be associated with plant growth, grain size and
flowering timein rice. In a study conducted by Iwamoto et al.
(2009), transgenic plants carrying full length Rdd 1cDNA driven
by constitutive promoter were produced in order to study the
role of Rdd1 and in - vivo function of Dof gene in plants. The two
types of transgenic plants were produced viz. RDD 1-S, transgenic
containing sense strand and RDD 1-AS with anti-sense strand.
Differences in the grain length, grain width and grain weight were
observed between RDD 1-AS and RDD 1-S plants when subjected
under varying photoperiod conditions [4]. When the grain sizes of
the two transgenic types of plants were compared with the wild
type plants, there was significant decrease in grain length, grain
width and decrease in 1000 grain weight in transgenics carrying
antisense gene (AS1=4.67 and AS2=4.54). In our study, we used
previously reverse RDD-1 primer and designed a new forward
primer giving an amplicon size of 620 bp. The three alleles obtained
in the present study with sizes of 600, 620 and 600’490 base pairs
were designated as A, B and C, respectively. Significant association
(Χ2=3.94, p<0.05) was observed between the grain length and
the allele A (600bp) for large grain length and allele B (620 bp)
associated with small grain length (Χ2=4.51, p<0.05). The allele A
for Rdd1 can be used in breeding for large grain length. In our study,
however, a set of 41 genotypes having large grain length ranging
from 8.27-10.31mm and small grain length of 5.48-7.52 mm were
used. This phenotypic variation is distinct from the WT genotype
Nipponbare (4.94mm) used by Iwamoto [4]. The allele A though
similar in size could be different due to nucleotide variation. In
order to rule out that possibility, sequencing of the allele from a
panel of genotypes showing large grain length will have to be done.
GS 5 (Grain size 5) QTL has been reported to be a positive
regulator of grain width and flanked by markers RM 593 and RM
574. Over-expression of GS5 promotes cell division resulting in
an increase of grain width [6]. Other genes/QTLs in rice are Grain
weight 7 (gw 7) reported to be associated with 1000 grain weight
located at chromosome 7 in population derived from O. sativa and
O. grandiglumis. Phenotypic variation of 13.3% was observed in
populations having allele for gw 7 from Caipo (Aluko, 2003). Grain
width 8 (gw 8) at chromosome 8 has been reported to have negative
effect on grain width with decrease in grain width in populations
resulting from separate crosses made between HJX 74 x Anmol
3 and HJX 74 x Basmati 370, with possible introgression of the
recessive allele from Anmol 3 and Basmati 370 [7,8]. No significant
association was observed for primers reported for Grain weight
8.1 (gw 8.1) [9] QTL in our panel of 41 genotypes. Grain thickness
7 (gt 7), a QTL for grain thickness with linked marker RM 478 at
chromosome 7¸ has been reported to be associated positively with
grain thickness in HG 101 which is a near isogenic line obtained
from O. sativa (Hwaseongbyeo) x O. grandiglumis. T [8].
he increased grain length in mm (5.53), grain width in mm
(3.08) and 1000 grain weight in grams (26.3) was observed for
the NILs as compared to the recipient parent, Hwaseongbyeo,
with grain length (5.03 mm), grain width (2.84mm) and 1000
grain weight (21.5g). In the present study, significant association
of primer RM 478 was observed and the allele A (205bp) was
significantly associated with low 1000 grain weight. The genotypes
used in the present study have highest 1000 grain weight ranging
from 20.16-36.04 grams and lowest ranging from 10.72-15.59
grams. Since grain yield is quantitative in nature, it is a complex
trait and the markers tagged with the genes/QTLs may not show
association in a diverse set of genotypes. Also the effect of individual
genic components to grain yield may be very low as grain yield is a
trait caused by cumulative action of many related components. Our
study however, suggests that the allele A (600 bp) from Rdd 1 can
be used in breeding programmes to enhance grain length in rice.
The genotypes viz. Hans raj, Pant dhan-18, Pant dhan-10, Narendra,
Pant dhan-12, Pant dhan-4, ARR-09, Shahsarang, IVT-ASG-2712,
IVT-ASG-2701, AVT-IASG-2602, AVT-IASG-2609 and IVT-ASG-2705
have been identified to be carrying this allele. These can be used
as a source of large grain length in future. The primers RM 478
for grain thickness and RM 574-2 for grain width can be used in
marker assisted breeding programmes. Allele A (205 bp) of RM 478
and allele B (240 bp) of RM 574-2 associated with low grain weight
and grain width, respectively can be used for rejection at seedling
stage in marker assisted breeding programme (Table 7). This study
also identified novel alleles (alleles distinct from those reported in
biparental programme); which could be significant in a larger panel
of rice genotypes (Figure 2-4).
Discussion
Phenotypic analysis for yield related traits
Phenotypic variation was observed for grain dimensions in
all the genotypes. The mean data for grain width (Figure 5), grain
length (Figure 6) and 1000 grain weight (Figure 7) was taken for all
the 41 genotypes used for allele mining study. The genotypes with
larger grain width (mm) were LR-1 (3.62), LR-71 (3.07), Pant dhan-
16 (2.85), IVT-2712 (2.79) and RWR-19 (2.77). Smaller grain width (mm)
was observed in Kalanamak (2.01), AVT-2607 (2.0), Hans raj
(1.97), AVT-2701 (1.94), IVT-2703 (1.904) and AVT-2609 (1.85).
The panel consisted of 39.02 % large (2.49-3.62), 51.22 % medium
(2.01-2.44) and 9.76 % small (1.85-1.97) for grain width (in mm)
trait. The genotypes with larger grain length (mm) were Hans
raj (10.31), Pant dhan-18 (9.83), Pant dhan-10 (9.7), Narendra
(9.63), Pant dhan-12 (9.53), RWR-125 (9.27), Pant dhan-4 (9.23)
and Govind (9.12). Smaller grain length (mm) was observed for
genotypes IVT-ASG-2708 (6.41), Kalanamak (6.35), IVT-ASG-2711
(6.15), IVT-ASG-2607 (5.92), IVT-ASG-2713 (5.71), IVT-ASG-2703
(5.69), LR-71 (5.59) and LR-77 (5.48). The genotype panel consisted
of 19.51 % large (9.18-10.31), 56.1 % medium (7.36-8.74) and
24.4 % small (5.48-6.82) for grain length (in mm). The genotypes
having the highest grain weight (g) were LR-1 (36.04), Pant dhan-
18 (27.97), Pant dhan-4 (26.39), LR-23 (25.32), RWR-125 (25.05),
Narendra (24.2), Pant dhan -12 (23.97) and Pant dhan-16 (23.39).
Lower grain weight (g) was observed for genotypes, Kalanamak
(12.17), LR-77 (12.14), AVT-IASG-2609 (12.00), IVT-ASG-2713
(11.96), AVT-IASG-2607 (10.74) and IVT-ASG-2703 (10.72) (Tables
8 & 9). The genotype panel consisted of 43.9 % high (23-36), 34.15
% medium (19.5-22.8) and 21.95 % small (17-19.45) for 1000
grain weight (g). The phenotypic variance observed for the 41 rice
genotypes were found to be significant at 5% level for grain width
(F=9.61) and grain length (F=6.1).
Figure 5: SSR profile of grain length generated by primer RDD1-2 in 41 rice genotypes. The numbers at the top refer to the
genomic DNA code as mentioned in Table 4.2. Alleles are indicated at the bottom. Allele A=approximately 600 bp; allele B=620
bp; B’C=620’ 490 bp; X=no amplification. 1kbp ladder is indicated at the right of the gel.
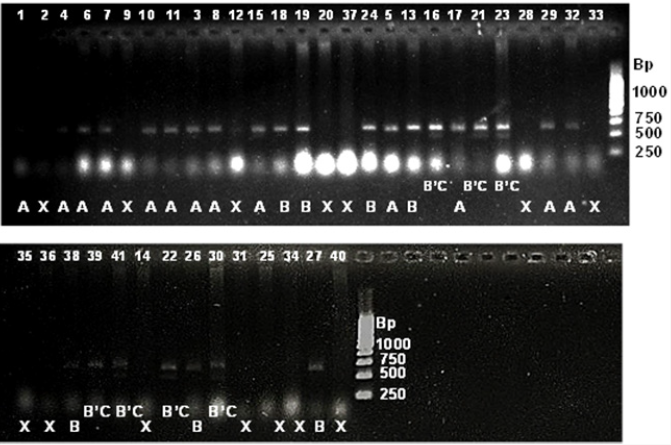
Figure 6: SSR profile of grain weight generated by primer RM 234 in 41 rice genotypes. The numbers at the top refer to the
genomic DNA code as mentioned in Table 4.2. Alleles are indicated at the bottom. Allele A=approximately 110 bp; allele B=125
bp; allele C=155; X=no amplification. 100 bpladder is indicated to the right of the gel.

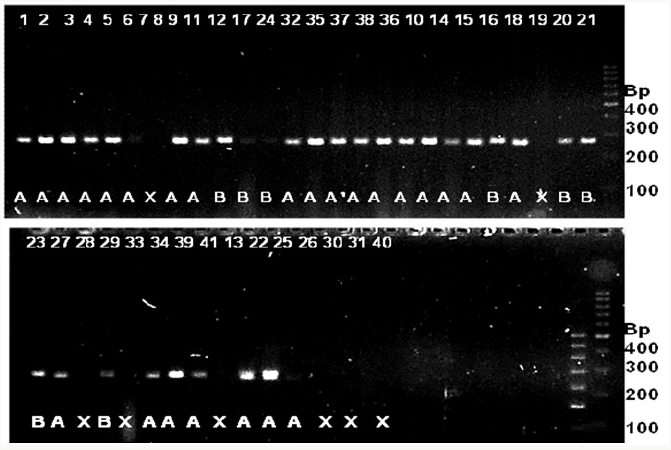
Figure 7: SSR profile of grain width generated by primer RM 502 in 41 rice genotypes. The numbers at the top refer to the
genomic DNA code as mentioned in Table 4.2. Alleles are indicated at the bottom. Allele A=approximately 255 bp; B=170 bp;
X=no amplification. 100 bpladder is indicated to the right of the gel.
Allele mining for genes/QTLs
Alleles are the alternative DNA sequences at the same physical
locus which may or may not result in different phenotypic traits.
Increasing demand of rice varieties with higher yield is making
scientists work for the identification of superior and novel alleles
to be used in breeding programmes. Allele mining is another
very important molecular approach towards identifying and
bringing together useful traits from diverse genetic sources
and introgression these useful alleles into a common recipient
promising cultivar lacking one or two superior alleles for some
agronomic traits. Molecular markers are effectively being used to
detect the variant alleles present in diverse genotypes or cultivars
which are phenotypically different from each other for the traits
in question. Microsatellite markers or SSR markers are PCR-based
markers that specifically bind to the complementary sequence of
the plant genomic DNA during PCR reaction. Through allele mining
novel genes could be identified from diverse sources and used in
improvement of crops for specific traits. Allele mining for grain
yield related traits in large number of genotypes/cultivars would
help in identifying new alleles of the genes known for the target
trait and their sources. It can also provide an insight into molecular
basis of novel trait variations and identify the nucleotide sequence
changes associated with superior alleles.
The quantitative trait locus (QTL) mapping has contributed to
a better understanding of the genetic basis of many agronomically
important traits such as grain yield. Grain yield is a quantitatively
inherited trait which is the result collectively contributed by several
individual grain traits. Of these, grain length, grain width and grain
weight are some of the important yield contributing grain traits in
rice. Today, a number of genes/QTLs associated with these traits
have been identified, sequenced, mapped and many associated
markers to these QTLs have been developed recently. In this study,
a set of 41 diverse genotypes including two accessions viz. RWR-
19 and RWR-125 from wild species O. nivarawere used for allele
mining using nine SSR markers tagged to reported genes/QTLs.
Out of the nine primers viz. RDD 1-2, RM 234, RM 478, RM 23201,
RM 502, RM 574-1, RM 574-2, RM 593-1 and RM 593-2 used in the
present study, RM 23201 was found to be monomorphic and the
rest eight primers were polymorphic.
Association between genotype and phenotype
Chi square test was conducted to study the association of the
markers with grain traits. Each of the alleles observed in the top 15
and bottom 15 genotypes were tested for the consistency of their
distribution with the whole population. Significant association
(Χ2=3.94, p=0.05) was observed for allele A in the 15 genotypes
having large grain length for primer RDD 1-2. Allele Bfor Rdd 1 gene
too showed significant association (Χ2=4.51, p=0.03). Allele A was
found to be associated with large grain length and allele B was found
to be associated with small grain length. Significant association
was observed for marker RM 478 with grain weight and for the
distribution of A allele in the bottom 15 genotypes (lowest grain
weight) with the distribution in the overall population (Χ2=5.08,
p=0.02) and for primer RM 574-2, distribution of B allele (Χ2=4.3,
p=0.08) in the bottom 15 genotypes (smallest grain width) was
found to be significant. Rdd 1(Rice dof daily fluctuations 1) Dof gene
in rice has been reported to be associated with plant growth, grain
size and flowering time in rice. In a study conducted by Iwamoto
et al. (2009), transgenic plants carrying full length Rdd 1 cDNA
driven by constitutive promoter were produced in order to study
the role of Rdd 1 and in vivo function of Dof gene in plants. The two
types of transgenic plants were produced viz. RDD 1-S, transgenic
containing sense strand and RDD 1-AS with anti-sense strand. Differences in the grain length, grain width and grain weight were
observed between RDD 1-AS and RDD 1-S plants when subjected
under varying photoperiod conditions [4]. When the grain sizes of
the two transgenic types of plants were compared with the wild
type plants, there was significant decrease in grain length, grain
width and decrease in 1000 grain weight in transgenics carrying
antisense gene (AS1=4.67 and AS2=4.54).
In our study, we used previously reverse RDD-1 primer
and designed a new forward primer giving an amplicon size of
620bp. The three alleles obtained in the present study with sizes
of 600, 620 and 600’490 base pairs were designated as A, B and
B’C, respectively. Significant association (Χ2=3.94, p< 0.05) was
observed between the grain length and the allele A (600bp) for
large grain length and allele B (620bp) associated with small grain
length (Χ2=4.51, p<0.05). The allele A for Rdd 1 can be used in
breeding for large grain length. In our study, however, a set of 41
genotypes having large grain length ranging from 8.27-10.31 mm
and small grain length of 5.48-7.52 mm were used. This phenotypic
variation is distinct from the WT genotype Nipponbare (4.94mm)
used by Iwamoto [4]. The allele A though similar in size could
be different due to nucleotide variation. In order to rule out that
possibility, sequencing of the allele from a panel of genotypes
showing large grain length will have to be done. GS 5 (Grain size
5) QTL has been reported to be a positive regulator of grain width
and flanked by markers RM 593 and RM 574. Over-expression of
GS 5 promotes cell division resulting in an increase of grain width
[6]. Other genes/QTLs in rice are Grain weight 7 (gw 7) reported to
be associated with 1000 grain weight located at chromosome 7 in
population derived from O. sativa and O. grandiglumis. Phenotypic
variation of 13.3% was observed in populations having allele for
gw 7 from Caipo [1]. Grain width 8 (gw 8) at chromosome 8 has
been reported to have negative effect on grain width with decrease
in grain width in populations resulting from separate crosses made
between HJX 74 x Anmol 3 and HJX 74 x Basmati 370, with possible
introgression of the recessive allele from Anmol 3 and Basmati 370
[7,8]. No significant association was observed for primers reported
for Grain weight 8.1 (gw 8.1) [9] QTL in our panel of 41 genotypes.
Grain thickness 7 (gt 7), a QTL for grain thickness with
linked marker RM 478 at chromosome 7¸ has been reported to
be associated positively with grain thickness in HG 101 which is
a near isogenic line obtained from O. sativa (Hwaseongbyeo) x
O. grandiglumis [8]. The increased grain length in mm (5.53),
grain width in mm (3.08) and 1000 grain weight in grams (26.3)
was observed for the NILs as compared to the recipient parent,
Hwaseongbyeo, with grain length (5.03mm), grain width (2.84mm)
and 1000 grain weight (21.5g). In the present study, significant
association of primer RM 478 was observed and the allele A
(205bp) was significantly associated with low 1000 grain weight.
The genotypes used in the present study have highest 1000 grain
weight ranging from 20.16-36.04 grams and lowest ranging from
10.72-15.59 grams. Since grain yield is quantitative in nature, it is
a complex trait and the markers tagged with the genes/QTLs may
not show association in a diverse set of genotypes. Also the effect
of individual genic components to grain yield may be very low as
grain yield is a trait caused by cumulative action of many related
components. Our study however, suggests that the allele A (600bp)
from Rdd 1 can be used in breeding programmes to enhance grain
length in rice. The genotypes viz. Hans raj, Pant dhan-18, Pant dhan-
10, Narendra, Pant dhan-12, Pant dhan-4, ARR-09, Shahsarang, IVTASG-
2712, IVT-ASG-2701, AVT-IASG-2602, AVT-IASG-2609 and IVTASG-
2705 have been identified to be carrying this allele. These can
be used as a source of large grain length in future. The primers RM
478 for grain thickness and RM 574-2 for grain width can be used
in marker assisted breeding programmes. Allele A (205bp) of RM
478 and allele B (240bp) of RM 574-2 associated with low grain
weight and grain width, respectively can be used for rejection at
seedling stage in marker assisted breeding programme. This study
also identified novel alleles (alleles distinct from those reported in
biparental programme); which could be significant in a larger panel
of rice genotypes (Tables 7-9).
Table 7: List of reported candidate genes/QTL, their associated markers, location, and associated grain traits used for allele mining
study.
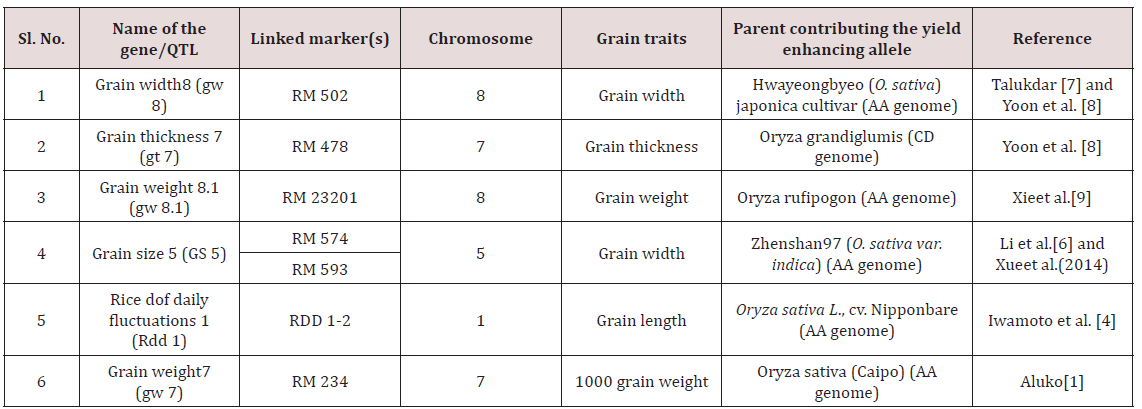
Table 8: Analysis of variance for grain width in rice genotypes.
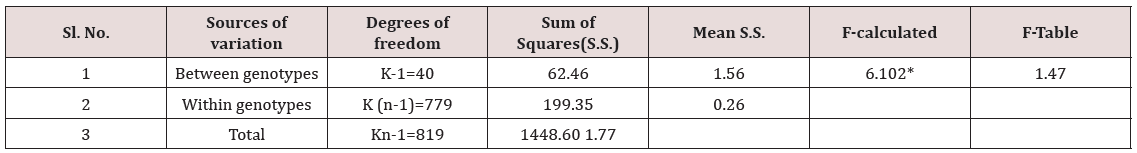
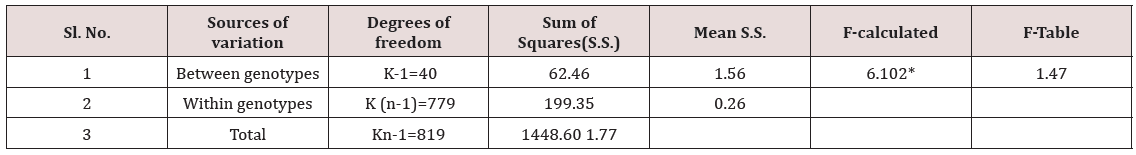
Table 9: Analysis of variance for grain length in rice genotypes.
Conclusion
From this present experiment, hybrid plants from inter-specific
crosses can be used for backcrossing in future plant breeding
programmes. Crosses can also be attempted for genotypes that
failed during this experiment. Primer RDD 1-2 can be used for
identifying alleles enhancing grain length from diverse genomic
sources for crop improvement. The allele A (600 bp) from Rdd
1 can be used in breeding programmes to enhance grain length
in rice. The genotypes viz. Hans raj, Pant dhan-18, Pant dhan-10,
Narendra, Pant dhan-12, Pant dhan-4, ARR-09, Shahsarang, IVTASG-
2712, IVT-ASG-2701, AVT-IASG-2602, AVT-IASG-2609 and
IVT-ASG-2705 identified carrying this allele can be used as a source
for large grain length in future. The primers RM 478 for grain
thickness and RM 574-2 for grain width can be used in marker
assisted breeding programmes. Allele A (205bp) of RM 478 and
allele B (240bp) of RM 574-2 associated with low grain weight and
grain width respectively, can be used for rejection at seedling stage
in marker assisted breeding programme. This study also identified
novel alleles (alleles distinct from those reported in biparental
programme); which could be significant in a larger panel of rice
genotypes. Read More Lupine Publishers Agriculture Journal Articles: