Lupine Publishers| Journal of Biomedical Engineering and Biosciences
Abstract
The use of seaweeds as functional foods and drugs is well known. Capsosiphon fulvescens (C. fulvescens) is a green sea alga for which beneficial health impacts have been reported even in diabetes. In the present study, three C. fulvescens constituents (chalinasterol, capsofulvesin A and capsofulvesin B) were evaluated for their molecular binding signature and interaction patterns against human aldose reductase as an antidiabetic target. The enzyme has been robustly implicated in the development of secondary complications of diabetes. The results generated here with the aid of in silico tools revealed a favourable binding affinity for the seawed constituents on the active site of aldose reductase. However, detailed observation shows no interference with the NADP binding pocket. The binding configuration of capsofulvesin A and capsofulvesin B were comparable to fidarestatas the compounds inserted one of their elongated aliphatic long chains with double bonds into the binding pocket. Nonetheless, subtle differences were sighted with the binding format of the aliphatic chain of capsofulvesin B resulting from a possible hydrophobic hinderance or influence within the binding pocket. Only one of the aliphatic chains of capsofulvesin A enjoyed hydrophobic interaction with residues Val47, Trp219 and Phe122 compared to the capsofulvesin B having hydrophobic interaction with Phe122 at the second chain while the other aliphatic chain formed hydrophobic bond with Trp20, Val47, Trp219 and Leu300. Meanwhile, the galactose unit of the two compounds displayed similar binding orientation and interaction with amino acid residues Ser22 and Trp20 at the active site. Chalinasterol, on the other hand, established hydrogen bond with the imidazole ring of His110 and indole ring of Trp111 respectively. The current study represents the first attempt to model the interaction of these ligands with aldose reductase and the unique binding patterns might explain the mechanism and potency of C. fulvescens in the management of diabetes and its complications.
Keywords: Capsosiphon fulvescens; Aldose reductase; Molecular interaction; Capsofulvesin A; Capsofulvesin B; Chalinasterol
Abbrevations: NADP: Nicotinamide Adenosine Diphosphate; 3D: Three Dimensional
Introduction
The human aldose reductase is the protein that catalyzes the rate-limiting step in polyol pathway which generates sorbitol. Due to elevated blood glucose level (hyperglycemia) in diabetic patients, metabolism of glucose by this enzyme in the pathway may lead to biochemical imbalances causing increased intraocular pressure [13]. Moreover, accumulation of sorbitol within the mesangial and proximal tubule cells can lead to alterations in cellular myoinositol level [2]. It can also lower Na+/K+-ATPase activity [3,4]. Each ofthese effects has inherent detrimental effect in diabetes. In addition, the conversion of glucose to sorbitol requires NADPH while oxidation of sorbitol enhances NADH concentration with a concomitant rapid imbalance in the cytoplasmic redox state [4]. Therefore, a reduction in the concentration of NADPH in the cytosolic will upset the NADPH+/NADP+ ratio. This may compromise the process which generates a reduced glutathione in oxidatively stressed cells [4,5]. Interestingly, aldose reductase inhibition can prevent or delay such numerous early alterations in hyperglycemic patient. Little wonder the enzyme has been validated as a therapeutic target in the management of diabetes and its complications.
Seaweed extracts have been reported for their diverse pharmacological and biological activities such as immunomodulating, anticoagulating, anticancer effects, etc [6- 8]. Capsosiphon fulvescens (C. fulvescens) commonly known as 'Maesaengi' is a green alga widely found in the coastal area of Koreaas well as along the coasts of North America and Europe [9,10]. The alga is generally eaten as food among the Koreans because of its unique flavor and soft texture as well as its acclaimed health-promoting properties. Studies have shown that it is rich in macronutrients like iron, potassium, micronutrient like iodine and vitamins (A and C) [11]. The seaweed is reportedly cultivated on a large-scale in both the laboratory and the field which enhances its economic value [12]. However, research efforts still continue till date to investigate its nutritional profile and potential therapeutic benefits [12-14]. A lot of studies carried out on C. fulvescens using in vitro and in vivo assays have unraveled the pharmacological potentials of the alga. When eaten, the seawee dis said to treat stomach disorders as well as in hangovers [11]. Extracts obtained from C. fulvescens was reported to inhibit platelet aggregation and reduce serum lipid level in ovariectomized animals [15]. The radical scavenging activities and ferric reducing ability of the seaweed extracts was reported by Cho et al. [16]. The presence of phenolic compounds and flavonoids in the sea plant was also reported and correlated with the antioxidant effectiveness of the plant. These compounds contribute to the capacity of C. fulvescens to improve lipid peroxidation rats orally fed high-carbohydrate and high-fat diet [17]. Son and coworkers observed the potent capacity of the seaweed to inhibit colonic aberrant Crypt Foci in experimental animals treated with azoxymethane [18]. The seaweed also reduces serum cholesterol levels in hypercholesterolemic animal model [17]. Furthermore, extracts of this alga is useful in developing natural herbicidal compounds.
Components of C. fulvescens have also been proved to be biologically active. For instance, a water-soluble polysaccharide obtained from this seaweed was reported to possess anticancer activity against various human cancer cell lines including prostate (PC-3), colon (HT-29), lung (A-549), and gastric cancer cell line (MK-N-45) [19,20]. Another report claimed that polysaccharides isolated from the alga can stimulate the growth of gastrointestinal cells [21]. A group of sulfated polysaccharides from C. fulvescens also shows immunomodulatory, anti-coagulant and hepatoprotective activities in vivo [10,22].
Kim and coworkers reported that C. fulvescens-derived glycoprotein had pro-apoptotic potential on human gastric carcinoma cells [19]. Similarly, a protein purified from the seaweed demonstrated excellent antioxidant property against 2,2-diphenyl- 1-picrylhydrazyl radical, hydroxyl radical, superoxide anion, and hydrogen peroxide (H2O2) in vitro [23]. The protein, which was obtained from the hydrophilic compartments of the seaweed, also alleviate impaired spatial memory induced by chronic ethanol exposure in vivo [24]. Another compound, pheophorbide A, from the seaweed inhibited endothelial dysfunction mediated by glycation end product formation [25].
Few reports are available in literature on the inhibitory effect of C. fulvescens and its components on enzymatic activity of proteins. Cho and colleague showed that extract from the alga can inhibit the enzymatic activity of alcohol dehydrogenase, glucosidase, elastase and angiotensin converting enzyme in vivo in a dose-dependent manner [9] while Yoo et al. [26] reveals the ability of the C. fulvescens to inhibit melanogenesis via downregulation of tyrosinase in B16 cells. These compounds also exert inhibitory effect on acetylcholinesterase as the target for their anti-neurodegenerative activity [27,28]. Although the antidiabetic potential of C. fulvescens and its constituents have been reported, there is dearth of evidence to show the precise molecular interaction with the target protein. Herein, an attempt was made to build the interaction model of antidiabetic components isolated from C. fulvescens with aldose reductase in silico towards unravelling the possible mechanisms involved in their inhibitory potential against the protein.
Material and Methods
In silico methods used in this study has been described previously [29].
Preparation of Ligands
Briefly, a total of five ligands were selected from the literature for docking study. Out of these compounds, three are natural compounds isolated from C. fulvescens while the other two are known aldose reductase inhibitors which were adopted as reference ligands. The chemical structure of these compounds was obtained from NCBI PubChem compound database (http://www. ncbi.nlm.nih.gov/pccompound) and prepared using Marvinsketch. Three-dimensional optimization of the ligand structures was done using Merck molecular force field (MMFF94) before use in docking studies. The ligands were saved as MOL SD (3D) format after optimization.
Selection and Preparation of Protein Structure
The starting three dimensional (3D) structure of the proteins used in this study was downloaded from the RCSB protein data bank (http://www.rcsb.org) with PDB ID: 1EF3. The macromolecule was found in complex with fidarestat and Nicotinamide Adenosine Diphosphate (NADP). All the co-crystallized water molecules found with the protein structure and fidarestat were deleted before molecular docking procedures.
Validation of Molecular Docking Procedure
One of the major ways of validating docking procedure is to accurately regenerate both the pose and the molecular interaction of the co-crystallized ligand on the crystallographically-determined protein structure. The ligand found at the binding site of the experimentally determined aldose reductase was deleted. The structure of the ligand (sdf format) was separately prepared using Marvin sketch as described above and re-docked into the enzyme active site. The molecular interaction, majorly hydrogen bond in this case, was compared to that of the x-ray diffraction crystal structure.
Molecular Docking
Autodock Vina 4.2 was used for molecular docking analysis and scoring [30,31]. The optimized ligand molecules were docked into the active site of refined model of aldose reductase. The rotatable bonds of the ligands were set to be free while the protein macromolecule was treated as a rigid structure. Grid box size was set at 100x100x100 whereas grid points were adjusted to -8.62, 40.82 and -7.18 A° (x, y, and z) to include all the amino acid residues at the binding site and permit. This gives enough space to enhance adequate ligand rotation and translation. However, the spacing between grid points was kept at 0.375 angstroms. After molecular docking experiment, the best results in terms of binding energy and pose were selected for analysis.
Data Analysis
All protein snapshots were taken using PYMOL while receptor- ligand molecular interaction was analysed on proteinsplus server [32].
Results and Discussion
Figure 1: 3D structure of aldose reductase in cartoon and surface representation respectively showing the binding site for inhibitor fidarestat (yellow stick) and NADPH (red stick) at the active site
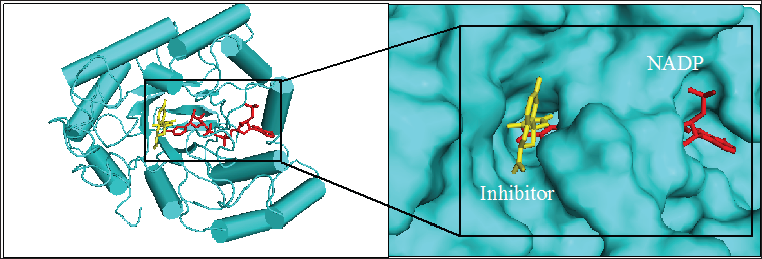
Figure 2: Chemical structure of the seaweed-derived compounds used as ligands in this study. (A)capsofulvesin A, (B) capsofulvesin B and (C) chalinasterol
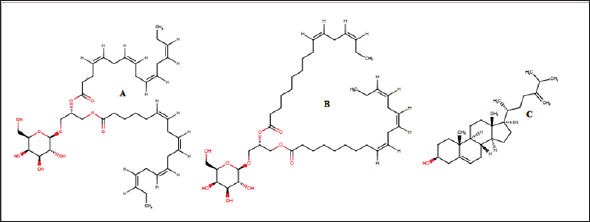
Figure 1 shows the structure of the protein, aldose reductase used as a target in the current study. Aldose reductase is an NADP- dependent enzyme which catalyzes the conversion of glucose to sorbitol in the polyol pathway. This pathway is essential because hyperglycemia, an occasion where blood glucose level becomes excessively high, can trigger an increased activity of aldose reductase which can lead to the accumulation of intracellular sorbitol. This effect has been implicated in the pathogenesis of diabetic complication such as retinopathy, neuropathy and nephropathy. Therefore, inhibition of the catalytic function of aldose reductase has been proposed as a promising therapeutic target in the prevention and treatment of type II diabetes and its associated it's severe degenerative complications [33]. As shown in Figure 1, the enzyme active site is connected to the NADP binding pocket. This provides a chance for a ligand to potentially be a competitor of either the substrate of the coenzyme. Some natural and synthetic inhibitors of this protein have been sought over the years. Most of the reported inhibitors of aldose reductase preferably bind to the substrate binding location rather than competing with NADPH [34]. For more accurate understanding of the aldose reductase- ligand compex in the present study, two known inhibitors of the protein (fidarestat [35] and inhibitor-IDD384 [34]) were employed to compare the interaction pattern. The 2D chemical structure of the alga-derived ligands are given in Figure 2. Chalinasterol is a cholesterol nucleus-containing compound and it is structurally different from capsofulvesin A and capsofulvesin B which are galactolipids comprising of double aliphatic long chains with double bonds. These ligands are naturally-occurring compounds isolated from pharmacologically-important seaweed (C. fulvescens) [28]. It is essential to emphasize that these bioactive compounds have earlier been reported to inhibit aldose reductase in vitro [36]. However, the mechanism of the inhibitory effect still remains uncler.
Figure 3: Analysis of the binding configuration of (A) chalinasterol, (B) capsofulvesin A and (C) capsofulvesin B. The binding pose for the compounds were compared against inhibitors of aldose reductase,fidarestat (green) and inhibitor- IDD384 (magenta). Areas with subtle differences in binding pose are identified in red spheres.
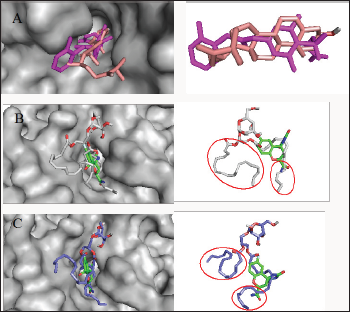
Chalinasterol shared a comparable binding pose with inhibitor- IDD384 (Figure 3). The molecular interaction as analyzed on proteinsplus server shows the contribution of both hydrophilic and hydrophobic bonds to the stability of the complex formed by the enzyme and C. fulvescens compounds (Figure 4-6). The bulky steroidal rings of chalinasterol was seen deeply buried within the fidarestat-binding pocket on aldose reductase where it is participated in hydrophobic bonds while the alkyl portion was exposed to the solvent-accessible region. Meanwhile, the inability of chalinasterol to completely block the active site may favor enzyme-substrate-chalinasterol complex formation as a mixed type of inhibition. As presented in Figure 4, two hydrogen bonds were established with amino acid residues His 110 and Trp111 respectively while Phe122, Trp219 and Trp20 participated in hydrophobic interaction with the ligand. The significance of hydrophobic bonds in the inhibition of aldose reductase cannot be overemphasized as it has been reported with many inhibitors of this protein including inhibitor IDD384.Inhibitory potential of chalinasterol against the carbohydrate-metabolizing enzyme was further proved by its binding affinity which is consistent with previous in vitro study that the compound possessed antidiabetic property [36,37]. The binding energy was relatively higher than that of the control ligands indicating a lower inhibitory activity for chalinasterol (Table 1). For capsofulvesin A and capsofulvesin B, the aromatic ring was placed at the solvent exposed region of the active site where it established hydrophilic interactions between its methoxyl moieties and the indole ring of Trp20 as well as the hydroxyl side chain of Ser22 (Figure 5). Possibly, the determining factor in the potency of these compounds as depicted in their IC50 is the binding configuration of the two long aliphatic chain. Capsofulvesin A has both of its side chain obstructing the entrance to the binding cavity while one of the aliphatic chains in capsofulvesin B was found moving away from the active site (Figure 6). This elongated side chain established a unique hydrophobic bond with residue Phe122. An efficient blockage of aldose reductase active site by capsofulvesin A may alter substrate access to the protein binding site, hence interfering with the enzymatic activity. Furthermore, the extra hydrophobic bond found with Phe122 in capsofulvesin B might have hindered the possible "swinging" of the aliphatic side chain from occupying the active site. It is also not impossible that these ligands interact with the enzyme-substrate complex in a mixed type of inhibition [36]. These mechanisms possibly underline the variations in the IC50 values previously obtained for these compounds. The binding energy of -7.9 kcal/mol estimated for capsofulvesin A compared to capsofulvesin B (-7.7 kcal/mol) suggests a higher affinity of the former to the protein. This result is in agreement with the previous in vitro report showing IC50 values of 52.53 |iM and 101.92 |iM for capsofulvesin A and capsofulvesin B respectively [36]. Although seaweeds have been investigated in diabetes and its complications, only scanty reports are available for C. fulvescens. Among the few studies, effect of extracts from the seaweed were observed for antidiabetic activity in vivo using streptozotocin-induced diabetic rats [37,38]. Therefore, this study provides insights into at least a part of the mechanisms responsible for the activity.
Table 1: Docking results showing binding energy and molecular interaction.
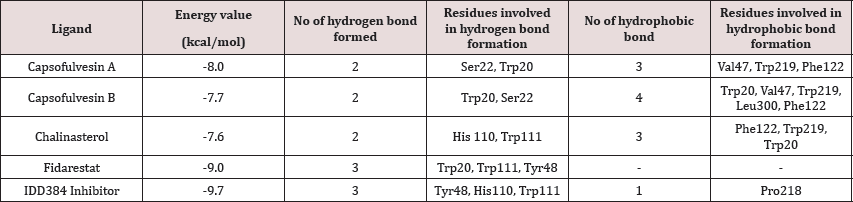
Figure 4: The binding pose and molecular the molecular interactions of chalinasterol with aldose reductase. Only key residues involved in hydrogen and hydrophobic interactions are shown as prepared on proteinsplus server.
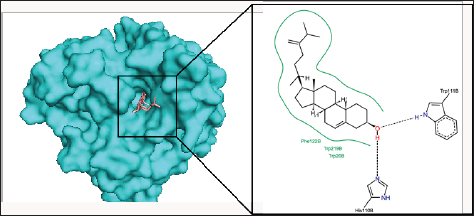
Figure 5: The binding pose and molecular the molecular interactions of capsofulvesin A with aldose reductase. Only key residues involved in hydrogen and hydrophobic interactions are shown as prepared on proteinsplus server.
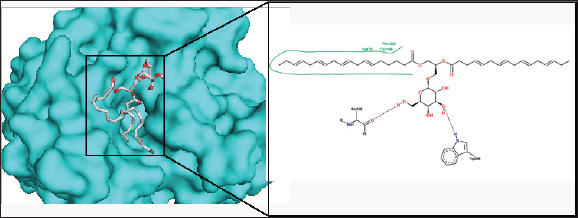
Figure 6: The binding pose and molecular the molecular interactions of capsofulvesin B with aldose reductase. Only key residues involved in hydrogen and hydrophobic interactions are shown as prepared on proteinsplus server.
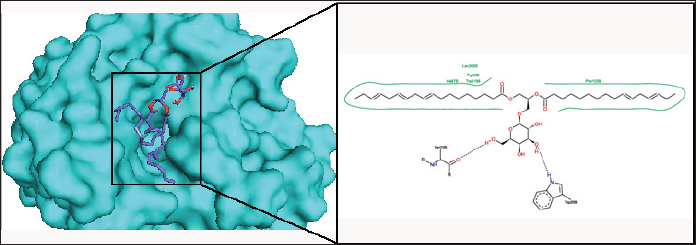
Conclusion
This study investigated three antidiabetic compounds from C. fulvescens (a well-known green sea algae) against a diabetes- related target (aldose reductase) for their possible binding and molecular interaction pattern to provide insights into their mechanism of inhibition. With binding configurations similar to that of known inhibitors of aldose reductase, the results of the current study divulge the alga-derived compounds as suitable inhibitors of the protein. They bind at the active site and established both hydrophilic and hydrophobic bonds with some amino acid residues around site. This indicates their potential to interfere with the catalytic function of the protein. However, it was clear that the compounds did not interact with the NADP binding pocket. Hence, they are not competitive with NADPH for their inhibitory activity against aldose reductase. The subtle contrasts in the binding mode of these compounds might be responsible for the differences in their IC50 values as previously reported in in vitro experiments. Taken together, the results of this study verify aldose reductase inhibitory activity of the C. fulvescens constituents and validate the antidiabetic property of the seaweed.
Read More About Lupine Publishers Journal of Biomedical Engineering and Biosciences Please Click on Below Link: https://biomedical-sciences-lupine-publishers.blogspot.com/

No comments:
Post a Comment
Note: only a member of this blog may post a comment.